🥁 Introduction
This notebook explores the capability of machine learning algorithms to distinguish between essays written by humans and those generated by Large Language models.
📐 Architecture
Using AWS ❝ Well-Architected Machine Learning Framework ❞ as a Guideline
Key Points:
- Design Philosophy: The architecture is aligned with the AWS 'Well-Architected Machine Learning' framework.
- Benefits:
- Ready to scale up going forward
- Ensures adherence to robust and efficient machine learning practices.
Importance:
- Best Practices: Aligns the project with industry standards in operational excellence, security, reliability, performance efficiency, and cost optimization.
- Future-Proofing: Even without current AWS implementation, this framework lays a strong foundation for potential AWS (and other cloud) integration.
Additional Resources:
- For a deeper understanding, visit AWS Well-Architected Machine Learning Framework.
🎮 Competition
https://www.kaggle.com/competitions/llm-detect-ai-generated-text
"Can you build a model to identify which essay was written by middle and high school students, and which was written using a large language model?"
"perhaps..."
📜 Hypothesis, Motivations and Objective
❈ Hypothesis: *Certain linguistic and structural patterns unique to AI-generated text can be identified and used for classification. We anticipate that our analysis will reveal distinct characteristics in AI-generated essays, enabling us to develop an effective classifier for this purpose.*
Motivations
- Learning and Challenge: Enhancing knowledge in Natural Language Processing (NLP) and staying intellectually active between jobs.
- Competition: https://www.kaggle.com/competitions/llm-detect-ai-generated-text
- Tool Development: Potential creation of a tool to differentiate between human and AI-generated content, useful across various fields.
- Educational Value: Serves as a practical introduction to production models in AI.
Objective
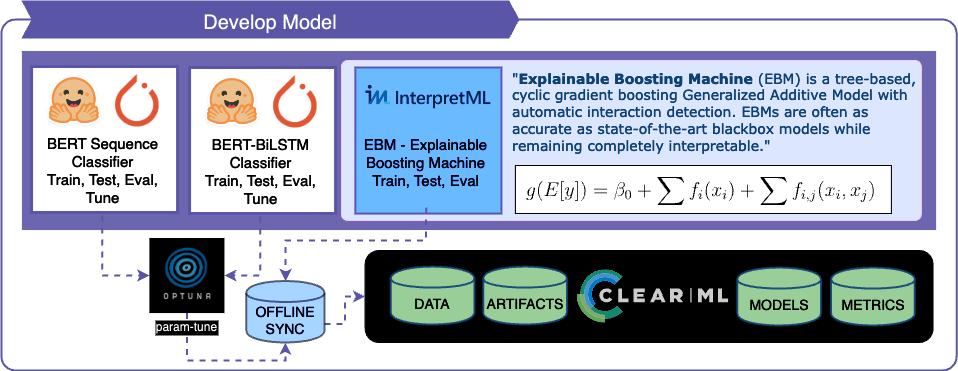
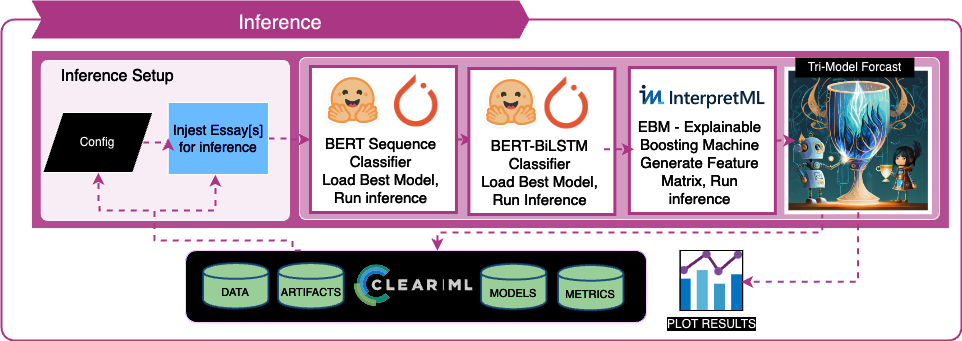
- Model Development: Building an ensemble model that combines two transformer models with an interpretable model like the Explainable Boosting Machine
- Challenges: Balancing effectiveness with interpretability.
- Approach:
- Developing an Explainable Boosting Machine (EBM) that uses custom features to compliment BERT's performance with better interpretability.
♪📕s
Data:
- https://www.kaggle.com/datasets/geraltrivia/llm-detect-gpt354-generated-and-rewritten-essays
- https://www.kaggle.com/datasets/thedrcat/daigt-v2-train-dataset
Models:
- https://www.kaggle.com/code/geraltrivia/ai-human-pytorchbertsequenceclassifier-model
- https://www.kaggle.com/code/geraltrivia/ai-human-pytorchcustombertclassifier-model
note: these are the two models in this notebook. Had to train in another, it's too expensive to run all at once.
Notebooks: This code and the ClearML pipelines including the Essay Generation are here https://github.com/mikewlange/ai-or-human.
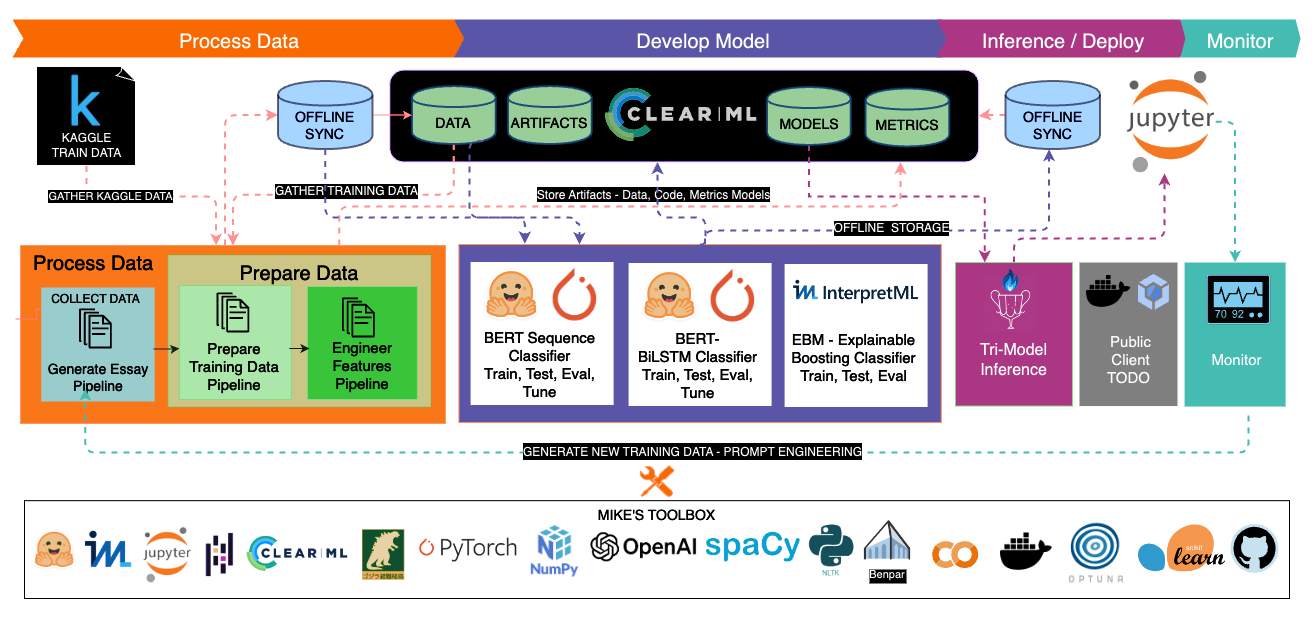
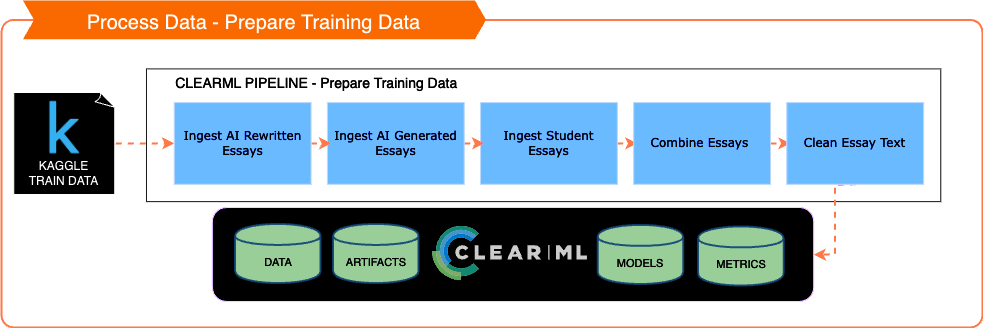
💡 ClearML Integration
"❤" - me
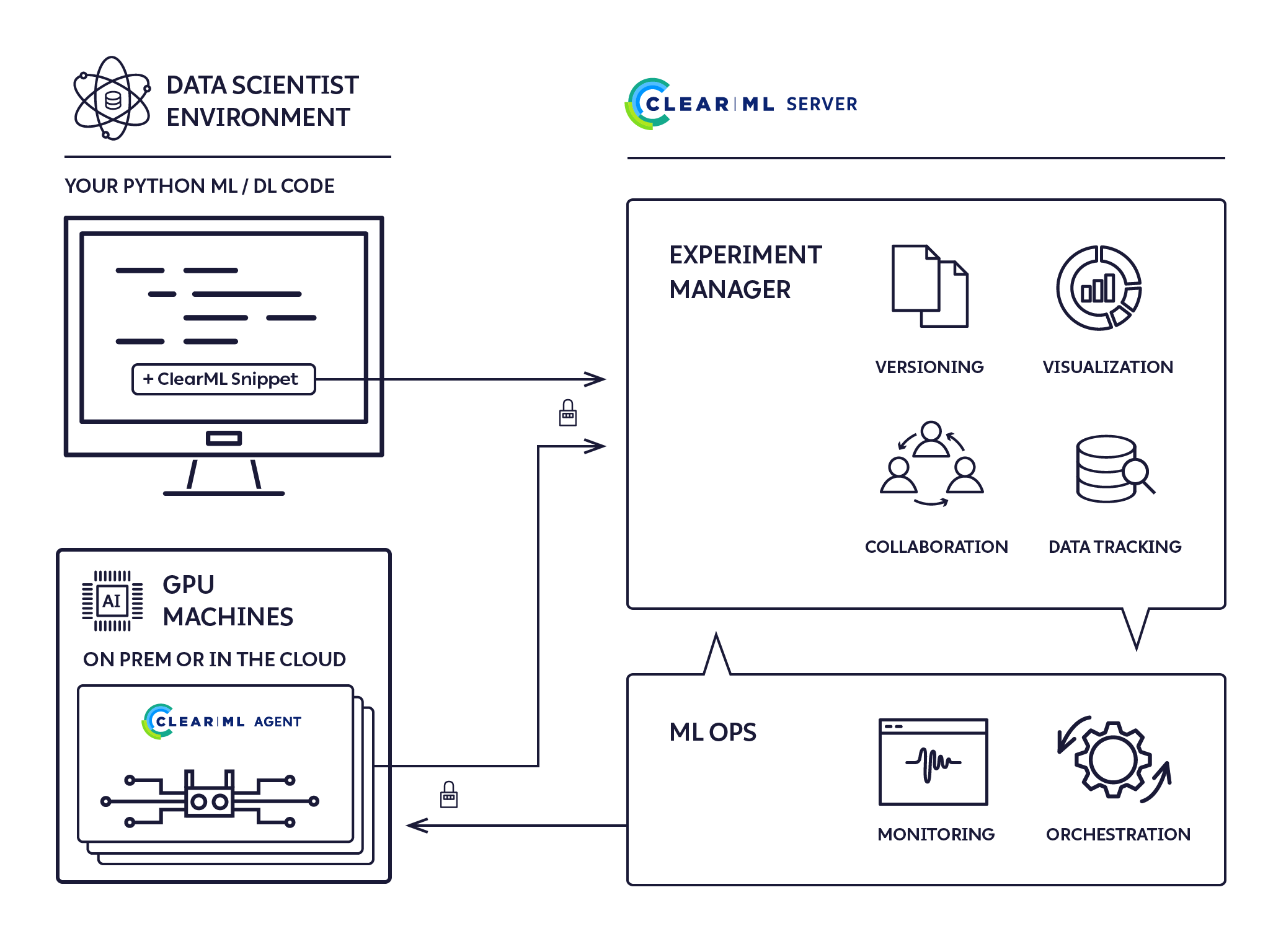
Strategic Choice and Benefits
- Purpose: ClearML integration to enhance workflow efficiency.
- Scalability and Efficiency: Focused on being prepared for scaling, even solo dev projects.
- Positive Experience: My experience with ClearML has been highly beneficial.
Implementation Details
- Pipelines Reference: In this project, all mentions of 'Pipelines' refer to ClearML pipelines.
- Resource Repository: A companion repository contains detailed code and examples for those interested in our ClearML implementation.
Impact of ClearML
- Operational Efficiency: ClearML has significantly improved the project's operational efficiency.
- Learning and Best Practices: It has also served as a platform for learning and applying best practices in ML operations, which is vital for scaling the project.
All the clearml related code is here: https://github.com/mikewlange/ai-or-human., but it's worth putting into the archtecture designs to help you see the whole picture.
✐ Setup for offline run
Kaggle Only
It's possible I'm doing this all wrong. To submit to a contest that disables the internet you neeed to add packages and other code/models that are not installed in the Kaggle docker image into a dataset (I.E storage) and use that as input to your notebook.
# turn off he internet and doing your pip installs, the packages you can't install, add to the libray var below
# TURN ON INTERNET FOR THIS
# creates a wheelhouse to add
#
# library = \
# '''
# textstat
# clearml
# sentence_transformers
# optuna
# interpret
# torchsummary
# empath
# benepar
# '''.lstrip('\n')
# with open('requirements.txt', 'w+') as f:
# f.write(library)
#!mkdir wheelhouse && pip download -r requirements.txt -d wheelhouse
# # Move requrements
# !mv requirements.txt wheelhouse/requirements.txt
## Zip it up and then you can download
# import os
# from zipfile import ZipFile
# dirName = "./"
# zipName = "packages.zip"
# # Create a ZipFile Object
# with ZipFile(zipName, 'w') as zipObj:
# # Iterate over all the files in directory
# for folderName, subfolders, filenames in os.walk(dirName):
# for filename in filenames:
# if (filename != zipName):
# # create complete filepath of file in directory
# filePath = os.path.join(folderName, filename)
# # Add file to zip
# zipObj.write(filePath)
# create a new dataset
# Take that zip file and add it to a dataset + button -> new dataset -> add all you need -> use as input here
#TURN OFF INTERNET
# wipe before any run. test. submission errors are no fun.
#!rm -rf /kaggle/working/*
# !cp -r /kaggle/input/pip-installs/wheelhouse /kaggle/working/
# !cp -r /kaggle/input/pip-installs/benepar_en3 /kaggle/working/
# !pip install --no-index --find-links=/kaggle/working/wheelhouse /kaggle/working/wheelhouse/benepar-0.2.0/benepar-0.2.0
# import sys
# sys.path.append("/kaggle/input/pip-installs/wheelhouse/sentence-transformers-2.2.2/sentence-transformers-2.2.2")
# import sentence_transformers
# sys.path.append("/kaggle/input/pip-installs/wheelhouse/empath-0.89/empath-0.89")
# from empath import Empath
# # Creating this in realtime just in case we have to add-remove.
# requirements = """
# textstat
# clearml
# optuna
# interpret
# torchsummary
# """
# with open('/kaggle/working/requirements.txt', 'w') as f:
# f.write(requirements)
# ## install
# !pip install -r /kaggle/working/requirements.txt --no-index --find-links /kaggle/input/pip-installs/wheelhouse
# !pip install --no-index --find-links=/kaggle/working/wheelhouse torchsummary
# # ## Prepare Benepar
# import sys
# import spacy
# import benepar
# import torchsummary
# # fixes ealier issue
# sys.path.insert(0, '/kaggle/working/')
# nlp = spacy.load('en_core_web_lg')
# nlp.add_pipe("benepar", config={"model": "benepar_en3"})
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import os
import random
import torch
import logging
from clearml.automation.controller import PipelineDecorator
from clearml import TaskTypes, PipelineController, StorageManager, Dataset, Task
from clearml import InputModel, OutputModel
from IPython.display import display
import ipywidgets as widgets
from tqdm import tqdm
import time
import pickle
import markdown
from bs4 import BeautifulSoup
import re
import nltk
from nltk.tokenize import word_tokenize
from nltk.stem import WordNetLemmatizer
torch.manual_seed(42)
if torch.cuda.is_available():
torch.cuda.manual_seed(42)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
# Set random seed for NumPy
np.random.seed(42)
# Set random seed for random module
random.seed(42)
#os.environ['OPENAI_API_KEY'] =
class CFG:
DEVICE = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
CLEAR_ML_TRAINING_DATASET_ID = 'e71bc7e41b114a549ac1eaf1dff43099'
CLEAR_ML_KAGGLE_TRAIN_DATA = '24596ea241c34c6eb5013152a6122e48'
CLEAR_ML_AI_GENERATED_ESSAYS = '593fff56e3784e4fbfa4bf82096b0127'
CLEAR_ML_AI_REWRITTEN_ESSAYS = '624315dd0e9b4314aa266654ebd71918'
DATA_ETL_STRATEGY = 1
TRAINING_DATA_COUNT = 50000
CLEARML_OFFLINE_MODE = False
CLEARML_ON = False
KAGGLE_INPUT = '/kaggle/input'
SCRATCH_PATH = 'scratch'
ARTIFACTS_PATH = 'artifacts'
TRANSFORMERS_PATH = 'benepar'
ENSAMBLE_STRATEGY = 2
KAGGLE_RUN = False
SUBMISSION_RUN = True
EXPLAIN_CODE=False
BERT_MODEL = 'bert-base-uncased'
EBM_ONLY = False
RETRAIN=True
cfg_dict = {key: value for key, value in CFG.__dict__.items() if not key.startswith('__')}
feature_list = list()
import clearml
class ClearMLTaskHandler:
def __init__(self, project_name, task_name, config=None):
self.task = self.get_or_create_task(project_name, task_name)
self.logger = None # Initialize logger attribute
self.setup_widget_logger()
if config:
self.set_config(config)
def get_or_create_task(self, project_name, task_name):
try:
tasks = []
if(CFG.CLEARML_OFFLINE_MODE):
Task.set_offline(offline_mode=True)
else:
tasks = Task.get_tasks(project_name=project_name, task_name=task_name)
if tasks:
if(tasks[0].get_status() == "created" and task[0].task_name == task_name):
task = tasks[0]
return task
else:
if(CFG.CLEARML_OFFLINE_MODE):
Task.set_offline(offline_mode=True)
task = Task.init(project_name=project_name, task_name=task_name)
return task
else:
if(CFG.CLEARML_OFFLINE_MODE):
Task.set_offline(offline_mode=True)
task = Task.init(project_name=project_name, task_name=task_name)
else:
task = Task.init(project_name=project_name, task_name=task_name)
return task
except Exception as e:
print(f"Error occurred while searching for existing task: {e}")
return None
def set_parameters(self, parameters):
"""
Set hyperparameters for the task.
:param parameters: Dictionary of parameters to set.
"""
self.task.set_parameters(parameters)
def set_config(self, config):
if isinstance(config, dict):
self.task.connect(config)
elif isinstance(config, argparse.Namespace):
self.task.connect(config.__dict__)
elif isinstance(config, (InputModel, OutputModel, type, object)):
self.task.connect_configuration(config)
else:
logging.warning("Unsupported configuration type")
def log_data(self, data, title):
self.task.get_logger()
if isinstance(data, np.ndarray):
self.task.get_logger().report_image(title, 'array', iteration=0, image=data)
elif isinstance(data, pd.DataFrame):
self.task.get_logger().report_table(title, 'dataframe', iteration=0, table_plot=data)
elif isinstance(data, str) and os.path.exists(data):
self.task.get_logger().report_artifact(title, artifact_object=data)
else:
self.task.get_logger().report_text(f"{title}: {data}")
def upload_artifact(self, name, artifact):
"""
Upload an artifact to the ClearML server.
:param name: Name of the artifact.
:param artifact: Artifact object or file path.
"""
self.task.upload_artifact(name, artifact_object=artifact)
def get_artifact(self, name):
"""
Retrieve an artifact from the ClearML server.
:param name: Name of the artifact to retrieve.
:return: Artifact object.
"""
return self.task.artifacts[name].get()
def setup_widget_logger(self):
handler = OutputWidgetHandler()
handler.setFormatter(logging.Formatter('%(asctime)s - [%(levelname)s] %(message)s'))
self.logger = logging.getLogger() # Create a new logger instance
self.logger.addHandler(handler)
self.logger.setLevel(logging.INFO)
# Just in case we can't use clearml in kaggle
class OutputWidgetHandler(logging.Handler):
def __init__(self, *args, **kwargs):
super(OutputWidgetHandler, self).__init__(*args, **kwargs)
layout = {'width': '100%', 'border': '1px solid black'}
self.out = widgets.Output(layout=layout)
def emit(self, record):
formatted_record = self.format(record)
new_output = {'name': 'stdout', 'output_type': 'stream', 'text': formatted_record+'\n'}
self.out.outputs = (new_output, ) + self.out.outputs
def show_logs(self):
display(self.out)
def clear_logs(self):
self.out.clear_output()
# Keeping this out for simpicity
def upload_dataset_from_dataframe(dataframe, new_dataset_name, dataset_project, description="", tags=[], file_name="dataset.pkl"):
from pathlib import Path
from clearml import Dataset
import pandas as pd
import logging
try:
print(dataframe.head())
file_path = Path(file_name)
pd.to_pickle(dataframe, file_path)
new_dataset = Dataset.create(new_dataset_name,dataset_project, description=description)
new_dataset.add_files(str(file_path))
if description:
new_dataset.set_description(description)
if tags:
new_dataset.add_tags(tags)
new_dataset.upload()
new_dataset.finalize()
return new_dataset
except Exception as e:
return logging.error(f"Error occurred while uploading dataset: {e}")
logging.basicConfig(level=logging.INFO)
logger = logging.getLogger(__name__)
( ͡° ͜ʖ ͡°) 𝖘𝖍𝖆K𝖊𝖘𝖕𝖊𝖆𝖗𝖊 𝖜𝖗o𝖙𝖊 𝖈o𝖉𝖊
⇟ Just toss explain_code(_i) into your cell, and voilà – it's like touching
a hot stove.
more info ⇟
[8] wikipedia
𝖘𝖍𝖆K𝖊𝖘𝖕𝖊𝖆𝖗𝖊?
- Deep Engagement: It encourages you to slow down and deeply engage with the content.
- Language Skill Enhancement: Instantly boosts your language skills. :)
- Programming and Language: Highlights the fact that programming is more about language than number crunching.
- Research Backing: Studies indicate that factors like fluid reasoning and language aptitude are crucial in understanding programming languages. [1]
- First Time: there is a good chance you've never read code explained like this.
𝖘𝖊T𝖚𝖕
- Config Section Update:
- Set your OpenAI API key:
os.environ['OPENAI_API_KEY'] = 'your_key'. - In the CFG object, set
EXPLAIN_CODE=True.
- Set your OpenAI API key:
- Code Explanation:
- Add
explain_code(_i)at the end of complex code cells.
- Add
- Execution:
- Run the cell and prepare for both enlightenment and a bit of humor.
from IPython.display import display, Markdown
import ipywidgets as widgets
from openai import OpenAI
client = OpenAI()
model = "gpt-4-1106-preview"
max_chars = 500
def query_openai_api(model, cell_contents, max_chars=500):
content = "" # Initialize the content variable
stream = client.chat.completions.create(
model=model,
response_format={"type": "text"},
messages=[
{"role": "system", "content": cell_contents },
{"role": "user", "content": "Analyze the code in the system message. Give a 3 sentence brief. Use a mild shakespearean tone based on a random charater from one of shakespears plays . Then, display the rest of explanation as a Markdown bullet list. Do not use a greetings. Focus on brevity and clarity. "}
],
max_tokens=max_chars,
temperature=0.7,
stream=True,
)
content = ""
display(Markdown(content))
for chunk in stream:
content += chunk.choices[0].delta.content or ""
if chunk.choices[0].finish_reason == "stop":
break
display(Markdown(content))
def explain_code(cell_contents):
loading_icon = widgets.HTML(value="")
loading_icon.layout.display = "none" # Hide the loading icon initially
output = widgets.Output()
def on_button_click(b):
with output:
loading_icon.layout.display = "block" # Show the loading icon
query_openai_api(model, cell_contents, max_chars)
loading_icon.layout.display = "none" # Hide the loading icon
button = widgets.Button(description="UNRAVEL MYSTERY", tooltip="Click to explain the code in this cell using gpt-4-1106-preview")
button.style.font_weight = "bold"
hbox = widgets.HBox([button, loading_icon])
button.background_color = "#05192D"
button.button_style= "primary"
button.style.width = "700px"
button.on_click(on_button_click)
display(hbox)
display(output)
if(CFG.EXPLAIN_CODE):
explain_code(_i)
🤣 𝕰x𝖆M𝖕l𝖊 ↑ 🤣
❝ Verily, the script before us, a modern parchment, doth invoke the spirits of computation to unravel the mysteries enfolded within its characters. Like Prospero's conjuring of airs and whispers, this code beckons forth answers from the aether, seeking knowledge with a sprite's swiftness. Yet not with charms or spells, but with the silent tongues of Python and OpenAI's grand oracle, it reveals its secrets. ❞
- The script creates an interactive widget within a Jupyter notebook that allows users to submit Python code for analysis.
- Upon clicking the "UNRAVEL MYSTERY" button, the code is sent to the OpenAI API, which uses the GPT-4 model to generate an explanation of the provided code snippet.
- The explanation is then displayed within the notebook, styled in Markdown for ease of reading.
- The function query_openai_api is responsible for communicating with the OpenAI API, handling the response, and formatting it as Markdown content.
- A loading icon is displayed while the API processes the request, providing visual feedback that the operation is in progress.
- The explain_code function sets up the interactive components, including the button and output area, and handles the button click event.
- The on_button_click function is the event handler that triggers the API call and manages the display of the loading icon and output.
Note on Functionality
- Current Limitation: Presently, it works on a 'load-and-reveal' basis - I want it to stream.
- Future Updates: Would like to get the streaming to work. :)
➠ Framework for Generating Essays
Quick overview. What are some ways students might use LLM's to conceal the origin of the essays?
- Simple Topic Essay Nothing fancy, simple instructions and the prompt
- Getting Creative Now we need to build a prompt that will fool the baseline model.
- Occasional grammatical errors: Introduce minor grammatical mistakes that a student might make under exam conditions or in a final draft, such as slight misuse of commas, or occasional awkward phrasing.
- Varying sentence structure: Use a mix of simple, compound, and complex sentences, with some variation in fluency to reflect a student's developing writing style.
- Personal touch: Include personal opinions, anecdotes, or hypothetical examples where appropriate, to give the essay a unique voice.
- Argument depth: While the essay should be well-researched and informed, the depth of argument might not reach the sophistication of a more experienced writer. Arguments should be sound but might lack the nuance a more advanced writer would include.
- Conclusion: Ensure the essay has a clear conclusion, but one that might not fully encapsulate all the complexities of the topic, as a student might struggle to tie all threads together neatly.
Remember, the goal is to create a piece that balances high-quality content with the authentic imperfections of a human student writer. The essay should be on the following topic: + prompt
- Rewrite Prompt That is where I would get caught. I love using AI to assist with rewrites and grammar checks.
Prompts were used from a competition dataset. https://www.kaggle.com/datasets/alejopaullier/daigt-external-dataset (original_moth)
The code for generating the essays is here: https://github.com/mikewlange/ai-or-human/blob/main/generate_essays_pipeline.ipynb
if(CFG.CLEARML_ON):
clearml_handler = ClearMLTaskHandler(
project_name='LLM-detect-ai-gen-text-LIVE/dev/notebook/preprocess',
task_name='Load Data and Generate Features'
)
clearml_handler.set_parameters({'etl_strategy': cfg_dict['DATA_ETL_STRATEGY'], 'train_data_count': cfg_dict['TRAINING_DATA_COUNT']})
clearml_handler.set_config(cfg_dict)
task = clearml_handler.task
def download_dataset_as_dataframe(dataset_id='593fff56e3784e4fbfa4bf82096b0127', file_name="ai_generated.pkl"):
import pandas as pd
# import Dataset from clearml
from clearml import Dataset
dataset = Dataset.get(dataset_id, only_completed=True)
cached_folder = dataset.get_local_copy()
for file_name in os.listdir(cached_folder):
if file_name.endswith('.pkl'):
file_path = os.path.join(cached_folder, file_name)
dataframe = pd.read_pickle(file_path)
return dataframe
raise FileNotFoundError("No PKL file found in the dataset.")
def download_dataset_as_dataframe_csv(dataset_id='593fff56e3784e4fbfa4bf82096b0127', file_name="ai_generated_essays.csv"):
import pandas as pd
# import Dataset from clearml
extension = file_name.split('.')[-1]
from clearml import Dataset
dataset = Dataset.get(dataset_id, only_completed=True)
cached_folder = dataset.get_local_copy()
for file_name in os.listdir(cached_folder):
if file_name.endswith(extension):
file_path = os.path.join(cached_folder, file_name)
dataframe = pd.read_csv(file_path)
return dataframe
kaggle_training_data = download_dataset_as_dataframe_csv(dataset_id=CFG.CLEAR_ML_KAGGLE_TRAIN_DATA,file_name="train_v2_drcat_02__final.csv")[['text','label','source']]
ai_generated_essays = download_dataset_as_dataframe(dataset_id=CFG.CLEAR_ML_AI_GENERATED_ESSAYS,file_name="ai_generated.pkl")[['text','label','source']]
ai_rewritten_essays = download_dataset_as_dataframe(dataset_id=CFG.CLEAR_ML_AI_REWRITTEN_ESSAYS,file_name="ai_rewritten_essays.pkl")[['text','label','source']]
random_kaggle_training_data = kaggle_training_data[kaggle_training_data['label'] == 1].sample(n=10000) # from kaggle dataset
random_generated_training_data = ai_generated_essays[ai_generated_essays['label'] == 1].sample(n=10000) # via the essay generation pipelint gpt3.5-4 essays written by AI and rewrittn
kaggle_training_student = kaggle_training_data[kaggle_training_data['label'] == 0].sample(n=12000)
random_kaggle_training_data = random_kaggle_training_data.dropna(subset=['text'])
random_generated_training_data = random_generated_training_data.dropna(subset=['text'])
kaggle_training_student = kaggle_training_student.dropna(subset=['text'])
combined_data = pd.concat([random_generated_training_data,random_kaggle_training_data, kaggle_training_student], ignore_index=True)
df_combined = combined_data.reset_index(drop=True)
df_combined.drop_duplicates(inplace=True)
df_essays = df_combined[['text', 'label', 'source']].copy()
sample = int(CFG.TRAINING_DATA_COUNT / 2)
df_label_0 = df_essays[df_essays['label'] == 0].sample(n=2000, random_state=42)
df_label_1 = df_essays[df_essays['label'] == 1].sample(n=2000, random_state=42) #<- example of data leakage
combined_df = pd.concat([df_label_1, df_label_0], ignore_index=True)
combined_df = combined_df.dropna()
df_essays = combined_df.reset_index(drop=True)
import plotly.graph_objects as go
import matplotlib.pyplot as plt
def plot_label_distribution(df_essays, plots, task=None):
if(plots == 1):
label_1_counts = df_essays[df_essays['label'] == 1].groupby('source').size()
label_0_counts = df_essays[df_essays['label'] == 0].groupby('source').size()
data=[
go.Bar(name='Label 0', x=label_0_counts.index, y=label_0_counts.values),
go.Bar(name='Label 1', x=label_1_counts.index, y=label_1_counts.values)
]
fig1 = go.Figure(data=data)
fig1.update_layout(
title='Counts of Label 0 and Label 1 per Source',
xaxis_title='Source',
yaxis_title='Count',
barmode='group'
)
if(CFG.CLEARML_ON):
task.get_logger().report_plotly(title="Counts of Label 0 and Label 1 per Source", series="data", figure=fig1)
# Show the chart using Plotly
fig1.show()
label_counts = df_essays['label'].value_counts().sort_index()
print("Label Counts:")
for label, count in label_counts.items():
print(f"Label {label}: {count}")
plt.bar(['0', '1'], label_counts.values)
plt.xlabel('label')
plt.ylabel('Count')
plt.title('Distribution of label Values in df_essays')
plt.show()
plot_label_distribution(df_essays,1)
if(CFG.EXPLAIN_CODE):
explain_code(_i)
INFO:matplotlib.category:Using categorical units to plot a list of strings that are all parsable as floats or dates. If these strings should be plotted as numbers, cast to the appropriate data type before plotting.
INFO:matplotlib.category:Using categorical units to plot a list of strings that are all parsable as floats or dates. If these strings should be plotted as numbers, cast to the appropriate data type before plotting.
Label Counts:
Label 0: 2000
Label 1: 2000
logging.basicConfig(level=logging.INFO, format='%(asctime)s - %(levelname)s - %(message)s')
def pipeline_preprocess_text(df):
PUNCTUATION_TO_RETAIN = '.?!,'
def preprocess_pipeline(text):
try:
# Remove markdown formatting
html = markdown.markdown(text)
text = BeautifulSoup(html, features="html.parser").get_text()
text = re.sub(r'[\n\r]+', ' ', text)
text = ' '.join(text.split())
text = re.sub(r'^(?:Task(?:\s*\d+)?\.?\s*)?', '', text)
text = re.sub('\n+', '', text)
text = re.sub(r'[A-Z]+_[A-Z]+', '', text)
punctuation_to_remove = r'[^\w\s' + re.escape(PUNCTUATION_TO_RETAIN) + ']'
text = re.sub(punctuation_to_remove, '', text)
tokens = word_tokenize(text)
lemmatizer = WordNetLemmatizer()
lemmatized_tokens = [lemmatizer.lemmatize(token) for token in tokens]
return ' '.join(lemmatized_tokens)
except Exception as e:
logging.error(f"Error in preprocess_pipeline: {e}")
return text
tqdm.pandas()
start_time = time.time()
df['text'] = df['text'].progress_apply(preprocess_pipeline)
end_time = time.time()
print(f"Preprocessing completed in {end_time - start_time:.2f} seconds")
return df
df_essays = pipeline_preprocess_text(df_essays)
if(CFG.CLEARML_ON):
plot_label_distribution(df_essays, 0, task=clearml_handler.task)
clearml_handler.task.upload_artifact(f'df_essays_train_preprocessed_{CFG.DATA_ETL_STRATEGY}', artifact_object=df_essays)
clearml_handler.task.get_logger().report_table(title='df_essays_train_preprocessed_',series='Train Essays Cleaned',
iteration=0,table_plot=df_essays)
if(CFG.EXPLAIN_CODE):
explain_code(_i)
0%| | 0/4000 [00:00<?, ?it/s]
100%|██████████| 4000/4000 [00:24<00:00, 160.03it/s]
Preprocessing completed in 25.01 seconds
⁖ 📲 Engineer Features
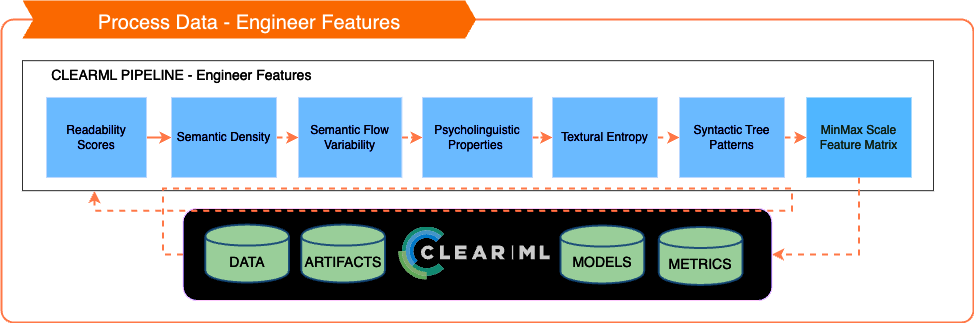
Key Analytical Areas
-
Readability Scores:
- Identifying unique patterns in AI vs. human-written essays.
- Analysis using scores like
Flesch-Kincaid Grade Level,Gunning Fog Index, etc.
-
Semantic Density:
- Understanding the concentration of meaning-bearing words in AI-generated vs. human text.
-
Semantic Flow Variability:
- Examining idea transitions between sentences in human and AI-generated texts.
-
Psycholinguistic Features:
- Using the LIWC tool for psychological and emotional content evaluation.
-
Textual Entropy:
- Measuring unpredictability or randomness, focusing on differences between AI and human content.
-
Syntactic Tree Patterns:
- Parsing essays to analyze syntactic tree patterns, especially structural tendencies in language models.
Ethical Considerations
- Content Bias: Ensuring to avoid generating discriminative content bias, focusing mainly on stat features.
- Potential Bias in Tools: Considering if tools like LIWC (empath) and the Readability Scores might introduce bias.
Note: These considerations are essential in maintaining the integrity and fairness of our analysis.
📊 Feature Distribution Statistics
Example:
Understanding Key Statistical Concepts
-
T-Test p-value:
- Purpose: Determines if differences between groups are statistically significant.
- Interpretation: A low p-value (< 0.05) suggests significant differences, challenging the null hypothesis.
-
Mann-Whitney U p-value:
- Usage: Ideal for non-normally distributed data, comparing two independent samples.
- Significance: Similar to the T-test, a lower p-value indicates notable differences between the groups.
-
Kruskal-Wallis p-value:
- Application: Used for comparing more than two independent samples.
- Meaning: A low p-value implies significant variance in at least one of the samples from the others.
-
Cohen's d:
- Function: Measures the standardized difference between two means.
- Values: Interpreted as small (0.2), medium (0.5), or large (0.8) effects.
-
Glass's delta:
- Comparison with Cohen's d: Similar in purpose but uses only the standard deviation of one group for normalization.
- Utility: Effective when the groups' standard deviations differ significantly.
Note on Sample Size and Statistical Tests
- Small Samples (Under 5000 Records): T-Test, Mann-Whitney U, and Kruskal-Wallis tests are effective.
- Large Samples (Over 5000 Records): Focus on effect sizes (Cohen's d and Glass's delta), as p-values will generally approach 0.
def plot_feature_distribution(df_essays, categories_to_plot, show_plot=True):
import plotly.graph_objects as go
import pandas as pd
import numpy as np
from scipy import stats
def cohens_d(x, y):
nx, ny = len(x), len(y)
dof = nx + ny - 2
return (np.mean(x) - np.mean(y)) / np.sqrt(((nx - 1) * np.std(x, ddof=1) ** 2 + (ny - 1) * np.std(y, ddof=1) ** 2) / dof)
def glass_delta(x, y):
return (np.mean(x) - np.mean(y)) / np.std(x, ddof=1)
for category in categories_to_plot:
df_filtered = df_essays[df_essays[category].astype(float) > 0]
generated_0 = df_filtered[df_filtered["label"] == 0][category].astype(float)
generated_1 = df_filtered[df_filtered["label"] == 1][category].astype(float)
# Statistical tests and effect size calculations
ttest_results = stats.ttest_ind(generated_0, generated_1, equal_var=False)
d_value = cohens_d(generated_0, generated_1)
delta_value = glass_delta(generated_0, generated_1)
u_statistic, p_value = stats.mannwhitneyu(generated_0, generated_1, alternative="two-sided")
k_statistic, p_value_k = stats.kruskal(generated_0, generated_1)
# Log scalar metrics
# logger.report_scalar(title="T-Test p-value", series=category, value=ttest_results.pvalue, iteration=0)
# logger.report_scalar(title="Cohen's d", series=category, value=d_value, iteration=0)
# logger.report_scalar(title="Glass's delta", series=category, value=delta_value, iteration=0)
annotations = (
f"<b>T-Test p-value:</b> {ttest_results.pvalue:.2e} <b> Mann-Whitney U p-value:</b> {p_value:.2e}<br>"
f"<b>Kruskal-Wallis p-value:</b> {p_value_k:.2e} <b> Cohen's d:</b>{d_value:.2f} <b> Glass's delta:</b>{delta_value:.2f}<br><br><br>"
)
fig = go.Figure()
data=[
go.Histogram(x=generated_0, name='Student', opacity=0.6),
go.Histogram(x=generated_1, name='AI', opacity=0.6)
]
fig.add_trace(data[0])
fig.add_trace(data[1])
condition = False
if (delta_value < -0.50) or (delta_value > 0.5):
condition = True
txt = 'title'
if condition:
title_text = f'<b>Distribution of {category.capitalize()}<span style="font-size: 30px; color: gold;">★</span></b>'
feature_list.append(category)
else:
title_text = f'<b>Distribution of {category.capitalize()}</b>'
fig.update_layout(
barmode='overlay',
title_text=title_text,
xaxis_title=f"{category.capitalize()}",
yaxis_title="Density",
annotations=[dict(
text=annotations,
x=.01,
y=-.25,
xref="paper",
yref="paper",
align="left",
showarrow=False,
bordercolor="#000000",
borderwidth=.3
)],
legend=dict(
orientation="v",
x=1.02,
y=1.0,
bgcolor="rgba(254, 255, 255, 0.5)",
bordercolor="#000000",
borderwidth=.3
),
margin=dict(l=100, r=100, b=100)
)
if(show_plot):
fig.show()
if(CFG.CLEARML_ON):
Task.current_task().get_logger().report_plotly(title=f"{category.capitalize()}", series="data", figure=fig)
Task.current_task().get_logger().report_single_value("T-Test p-value: ",ttest_results.pvalue)
Task.current_task().get_logger().report_single_value("Mann-Whitney U p-value: ",p_value)
Task.current_task().get_logger().report_single_value("Cohen's d: ",d_value)
Task.current_task().get_logger().report_single_value("Glass's delta:",delta_value)
⁖ 📲 Features
ƒ(①) Readability Scores
➠ Flesch-Kincaid Grade Level
This test gives a U.S. school grade level; for example, a score of 8 means that an eighth grader can understand the document. The lower the score, the easier it is to read the document. The formula for the Flesch-Kincaid Grade Level (FKGL) is:
$ FKGL = 0.39 \left( \frac{\text{total words}}{\text{total sentences}} \right) + 11.8 \left( \frac{\text{total syllables}}{\text{total words}} \right) - 15.59 $
Source: Wikipedia
➠ Gunning Fog Index
The Gunning Fog Index is a readability test designed to estimate the years of formal education a person needs to understand a text on the first reading. The index uses the average sentence length (i.e., the number of words divided by the number of sentences) and the percentage of complex words (words with three or more syllables) to calculate the score. The higher the score, the more difficult the text is to understand.
$ GunningFog = 0.4 \left( \frac{\text{words}}{\text{sentences}} + 100 \left( \frac{\text{complex words}}{\text{words}} \right) \right) $
In this formula:
- "Words" is the total number of words in the text.
- "Sentences" is the total number of sentences in the text.
- "Complex words" are words with three or more syllables, not including proper nouns, familiar jargon or compound words, or common suffixes such as -es, -ed, or -ing as a syllable.
The Gunning Fog Index is particularly useful for ensuring that texts such as technical reports, business communications, and journalistic works are clear and understandable for the intended audience.
Source: Wikipedia
➠ Coleman-Liau Index
The Coleman-Liau Index is a readability metric that estimates the U.S. grade level needed to comprehend a text. Unlike other readability formulas, it relies on characters instead of syllables per word, which can be advantageous for processing efficiency. The index is calculated using the average number of letters per 100 words and the average number of sentences per 100 words.
$ CLI = 0.0588 \times L - 0.296 \times S - 15.8 $
Where:
- L is the average number of letters per 100 words.
- S is the average number of sentences per 100 words.
Source: Wikipedia
➠ SMOG Index
The SMOG (Simple Measure of Gobbledygook) Index is a measure of readability that estimates the years of education needed to understand a piece of writing. It is calculated using the number of polysyllable words and the number of sentences. The SMOG Index is considered accurate for texts intended for consumers.
$ SMOG = 1.043 \times \sqrt{M \times \frac{30}{S}} + 3.1291 $
- M is the number of polysyllable words (words with three or more syllables).
- S is the number of sentences.
Source: Wikipedia
➠ Automated Readability Index (ARI)
The Automated Readability Index is a readability test designed to gauge the understandability of a text. The formula outputs a number that approximates the grade level needed to comprehend the text. The ARI uses character counts, which makes it suitable for texts with a standard character-per-word ratio.
$ ARI = 4.71 \times \left( \frac{\text{characters}}{\text{words}} \right) + 0.5 \times \left( \frac{\text{words}}{\text{sentences}} \right) - 21.43 $
Where:
- The number of characters is divided by the number of words.
- The number of words is divided by the number of sentences.
Source: wikipedia
➠ Dale-Chall Readability Score
The Dale-Chall Readability Score is unique in that it uses a list of words that are familiar to fourth-grade American students. The score indicates how many years of schooling someone would need to understand the text. If the text contains more than 5% difficult words (words not on the Dale-Chall familiar words list), a penalty is added to the score.
$ DaleChall = 0.1579 \times \left( \frac{\text{difficult words}}{\text{total words}} \times 100 \right) + 0.0496 \times \left( \frac{\text{total words}}{\text{sentences}} \right) $
$ \text{If difficult words} > 5\%: DaleChall = DaleChall + 3.6365 $
"Difficult words" are those not on the Dale-Chall list of familiar words.
Source: wikipedia
logging.basicConfig(level=logging.INFO)
logger = logging.getLogger(__name__)
tqdm.pandas()
# Start time
start_time = time.time()
#@PipelineDecorator.component(return_values=["df"], name='Readability Scores', task_type=TaskTypes.data_processing)
def process_readability_scores(df):
import logging
import textstat
try:
df['flesch_kincaid_grade'] = df['text'].progress_apply(textstat.flesch_kincaid_grade)
df['gunning_fog'] = df['text'].progress_apply(textstat.gunning_fog)
df['coleman_liau_index'] = df['text'].progress_apply(textstat.coleman_liau_index)
df['smog_index'] = df['text'].progress_apply(textstat.smog_index)
df['ari'] = df['text'].progress_apply(textstat.automated_readability_index)
df['dale_chall'] = df['text'].progress_apply(textstat.dale_chall_readability_score)
return df
except Exception as e:
logger.error(f"Error in process_readability_scores: {e}")
raise
%timeit
df_essays = process_readability_scores(df_essays)
end_time = time.time()
duration = end_time - start_time
print(f"Process completed in {duration:.2f} seconds")
0%| | 0/4000 [00:00<?, ?it/s]
100%|██████████| 4000/4000 [00:04<00:00, 996.32it/s]
100%|██████████| 4000/4000 [00:03<00:00, 1116.60it/s]
100%|██████████| 4000/4000 [00:00<00:00, 4238.36it/s]
100%|██████████| 4000/4000 [00:03<00:00, 1005.83it/s]
100%|██████████| 4000/4000 [00:00<00:00, 4843.97it/s]
100%|██████████| 4000/4000 [00:03<00:00, 1072.18it/s]
Process completed in 17.10 seconds
categories_to_plot = [
'flesch_kincaid_grade', 'gunning_fog', 'coleman_liau_index', 'smog_index', 'ari', 'dale_chall'
]
condition = True # Set the condition here
plot_feature_distribution(df_essays, categories_to_plot,True)
ƒ(②) Semantic Density
Calculating Semantic Density: The function calculate_semantic_density
computes this metric by determining the ratio of meaning-bearing words (identified by tags in
mb_tags) to the total word count. A higher semantic density indicates a text that efficiently
uses words with substantial meaning.
import nltk
from nltk.tokenize import word_tokenize
import string
# Start time
start_time = time.time()
def process_semantic_density(df):
# Configure logging
logging.basicConfig(level=logging.INFO, format='%(asctime)s - %(levelname)s - %(message)s')
def get_meaning_bearing_tags():
return {'NN', 'NNS', 'NNP', 'NNPS', 'VB', 'VBD', 'VBG', 'VBN', 'VBP', 'VBZ', 'JJ', 'JJR', 'JJS', 'RB', 'RBR', 'RBS'}
def tokenize_text(text):
try:
return word_tokenize(text.lower())
except TypeError as e:
logging.error(f"Error tokenizing text: {e}")
return []
def tag_words(words):
try:
return nltk.pos_tag(words)
except Exception as e:
logging.error(f"Error tagging words: {e}")
return []
def filter_words(tokens):
return [token for token in tokens if token.isalpha() or token in string.punctuation]
mb_tags = get_meaning_bearing_tags()
def process_row(text):
tokens = tokenize_text(text)
words = filter_words(tokens)
tagged = tag_words(words)
mb_words = [word for word, tag in tagged if tag in mb_tags]
full_sentence = " ".join(word + "/" + tag for word, tag in tagged)
density = len(mb_words) / len(words) if words else 0
return density, full_sentence
# Vectorized operations for DataFrame
df[['semantic_density', 'text_tagged_nltk']] = df['text'].progress_apply(lambda x: pd.Series(process_row(x)))
return df
%timeit
# run
df_essays = process_semantic_density(df_essays)
end_time = time.time()
# Calculate duration
duration = end_time - start_time
print(f"Process completed in {duration:.2f} seconds")
if(CFG.EXPLAIN_CODE):
explain_code(_i)
0%| | 0/4000 [00:00<?, ?it/s]
100%|██████████| 4000/4000 [01:22<00:00, 48.31it/s]
Process completed in 82.80 seconds
categories_to_plot = [
'semantic_density'
]
condition = True # Set the condition here
plot_feature_distribution(df_essays, categories_to_plot,True)
ƒ(⓷) Semantic Flow Variability
The model's approach, based on contrastive learning, is key to its effectiveness. It excels in distinguishing sentence pairs from random samples, aligning closely with the study's objective to analyze semantic flow.
import concurrent.futures
import logging
import nltk
import numpy as np
from sentence_transformers import SentenceTransformer
import torch
from tqdm import tqdm
tqdm.pandas()
import time
# Start time
start_time = time.time()
def process_semantic_flow_variability(df):
logging.basicConfig(level=logging.INFO, format='%(asctime)s - %(levelname)s - %(message)s')
logger = logging.getLogger(__name__)
# Load a pre-trained sentence transformer model
model_MiniLM = 'all-MiniLM-L6-v2' #https://huggingface.co/sentence-transformers/all-MiniLM-L6-v2
try:
model = SentenceTransformer(model_MiniLM)
except Exception as e:
logger.error(f"Error loading the sentence transformer model: {e}")
model = None
def cosine_similarity(v1, v2):
return torch.dot(v1, v2) / (torch.norm(v1) * torch.norm(v2))
def semantic_flow_variability(text):
if not model:
logger.error("Model not loaded. Cannot compute semantic flow variability.")
return np.nan
try:
sentences = nltk.sent_tokenize(text)
if len(sentences) < 2:
logger.info("Not enough sentences for variability calculation.")
return 0
sentence_embeddings = model.encode(sentences, convert_to_tensor=True, show_progress_bar=False)
# Calculate cosine similarity between consecutive sentences
similarities = [cosine_similarity(sentence_embeddings[i], sentence_embeddings[i+1])
for i in range(len(sentence_embeddings)-1)]
return torch.std(torch.stack(similarities)).item()
except Exception as e:
logger.error(f"Error calculating semantic flow variability: {e}")
return np.nan
if df is not None and 'text' in df:
# with concurrent.futures.ThreadPoolExecutor() as executor:
df['semantic_flow_variability'] = df['text'].progress_apply(semantic_flow_variability)
else:
logger.error("Invalid DataFrame or missing 'text' column.")
return df
%timeit
df_essays = process_semantic_flow_variability(df_essays)
end_time = time.time()
# Calculate duration
duration = end_time - start_time
print(f"Process completed in {duration:.2f} seconds")
# Explain the code in the cell. Add this line to each cell
if(CFG.EXPLAIN_CODE):
explain_code(_i)
INFO:sentence_transformers.SentenceTransformer:Load pretrained SentenceTransformer: all-MiniLM-L6-v2
INFO:sentence_transformers.SentenceTransformer:Use pytorch device: cpu
20%|██ | 807/4000 [01:03<04:06, 12.94it/s]INFO:__main__:Not enough sentences for variability calculation.
77%|███████▋ | 3099/4000 [04:01<01:01, 14.60it/s]INFO:__main__:Not enough sentences for variability calculation.
87%|████████▋ | 3491/4000 [04:24<00:36, 13.97it/s]INFO:__main__:Not enough sentences for variability calculation.
100%|██████████| 4000/4000 [04:55<00:00, 13.52it/s]
Process completed in 296.17 seconds
categories_to_plot = [
'semantic_flow_variability'
]
condition = True # Set the condition here
plot_feature_distribution(df_essays, categories_to_plot,True)
ƒ(⓸) Psycholinguistic Features
Psycholinguistic Features encompass the linguistic and psychological characteristics evident in speech and writing. These features provide insights into the writer's or speaker's psychological state, cognitive processes, and social dynamics. Analysis in this domain often involves scrutinizing word choice, sentence structure, and language patterns to deduce emotions, attitudes, and personality traits.
The Linguistic Inquiry and Word Count (LIWC) [3] is a renowned computerized text analysis tool that categorizes words into psychologically meaningful groups. It assesses various aspects of a text, including emotional tone, cognitive processes, and structural elements, covering categories like positive and negative emotions, cognitive mechanisms, and more.
While LIWC is typically accessible through purchase or licensing, this project will employ Empath, an open-source alternative to LIWC, to conduct similar analyses.
from empath import Empath
import pandas as pd
import logging
# Initialize logging
logging.basicConfig(level=logging.INFO)
logger = logging.getLogger(__name__)
# Create an Empath object
lexicon = Empath()
def empath_analysis(text):
try:
# Analyze the text with Empath and return normalized category scores
analysis = lexicon.analyze(text, normalize=True)
return analysis
except Exception as e:
# Log an error message if an exception occurs
logger.error(f"Error during Empath analysis: {e}")
# Return None or an empty dictionary to indicate failure
return {}
def apply_empath_analysis(df, text_column='text'):
"""
Apply Empath analysis to a column in a DataFrame and expand the results into separate columns.
"""
try:
df['empath_analysis'] = df[text_column].apply(empath_analysis)
empath_columns = df['empath_analysis'].apply(pd.Series)
df = pd.concat([df, empath_columns], axis=1)
df.drop(columns=['empath_analysis'], inplace=True)
return df
except Exception as e:
# Log an error message if an exception occurs
logger.error(f"Error applying Empath analysis to DataFrame: {e}")
# Return the original DataFrame to avoid data loss
return df
df_essays = apply_empath_analysis(df_essays)
# Explain the code in the cell. Add this line to each cell
if(CFG.EXPLAIN_CODE):
explain_code(_i)
columns_to_scale = ['help','office','dance','money','wedding','domestic_work','sleep','medical_emergency','cold','hate','cheerfulness','aggression','occupation','envy','anticipation','family','vacation','crime','attractive','masculine','prison','health','pride','dispute','nervousness','government','weakness','horror','swearing_terms','leisure','suffering','royalty','wealthy','tourism','furniture','school','magic','beach','journalism','morning','banking','social_media','exercise','night','kill','blue_collar_job','art','ridicule','play','computer','college','optimism','stealing','real_estate','home','divine','sexual','fear','irritability','superhero','business','driving','pet','childish','cooking','exasperation','religion','hipster','internet','surprise','reading','worship','leader','independence','movement','body','noise','eating','medieval','zest','confusion','water','sports','death','healing','legend','heroic','celebration','restaurant','violence','programming','dominant_heirarchical','military','neglect','swimming','exotic','love','hiking','communication','hearing','order','sympathy','hygiene','weather','anonymity','trust','ancient','deception','fabric','air_travel','fight','dominant_personality','music','vehicle','politeness','toy','farming','meeting','war','speaking','listen','urban','shopping','disgust','fire','tool','phone','gain','sound','injury','sailing','rage','science','work','appearance','valuable','warmth','youth','sadness','fun','emotional','joy','affection','traveling','fashion','ugliness','lust','shame','torment','economics','anger','politics','ship','clothing','car','strength','technology','breaking','shape_and_size','power','white_collar_job','animal','party','terrorism','smell','disappointment','poor','plant','pain','beauty','timidity','philosophy','negotiate','negative_emotion','cleaning','messaging','competing','law','friends','payment','achievement','alcohol','liquid','feminine','weapon','children','monster','ocean','giving','contentment','writing','rural','positive_emotion','musical']
plot_feature_distribution(df_essays, columns_to_scale,False) # too many, no need to view all.
ƒ(⓹) Textual Entropy
The standard method for calculating entropy is outlined below, which evaluates the unpredictability of each character or word based on its frequency. This approach is encapsulated in the formula for Shannon Entropy:
$\begin{aligned} H(T) &= -\sum_{i=1}^{n} p(x_i) \log_2 p(x_i) &&\quad\text{(Shannon Entropy)} \\ \end{aligned}$
Shannon Entropy quantifies the level of information disorder or randomness, providing a mathematical framework to assess text complexity.
import numpy as np
from collections import Counter
import logging
from tqdm import tqdm
tqdm.pandas()
import time
# Start time
start_time = time.time()
# Configure logging
logging.basicConfig(level=logging.INFO, format="%(asctime)s - %(levelname)s - %(message)s")
logger = logging.getLogger(__name__)
def calculate_entropy(text):
"""
Calculate the Shannon entropy of a text string.
Entropy is calculated by first determining the frequency distribution
of the characters in the text, and then using these frequencies to
calculate the probabilities of each character. The Shannon entropy
is the negative sum of the product of probabilities and their log2 values.
Args:
text (str): The text string to calculate entropy for.
Returns:
float: The calculated entropy of the text, or 0 if text is empty/non-string.
None: In case of an exception during calculation.
"""
try:
if not text or not isinstance(text, str):
logger.warning("Text is empty or not a string.")
return 0
# Calculating frequency distribution and probabilities
freq_dist = Counter(text)
probs = [freq / len(text) for freq in freq_dist.values()]
# Calculate entropy, avoiding log2(0)
entropy = -sum(p * np.log2(p) for p in probs if p > 0)
return entropy
except Exception as e:
logger.error(f"Error calculating entropy: {e}")
return None
%timeit
try:
df_essays["textual_entropy"] = df_essays["text"].progress_apply(calculate_entropy)
end_time = time.time()
duration = end_time - start_time
print(f"Process completed in {duration:.2f} seconds")
except Exception as e:
logger.error(f"Error applying entropy calculation to DataFrame: {e}")
end_time = time.time()
duration = end_time - start_time
print(f"Process completed in {duration:.2f} seconds")
if(CFG.EXPLAIN_CODE):
explain_code(_i)
100%|██████████| 4000/4000 [00:00<00:00, 6629.02it/s]
Process completed in 0.61 seconds
categories_to_plot = ['textual_entropy']
plot_feature_distribution(df_essays, categories_to_plot,True)
ƒ(⓺) Syntactic Tree Patterns
Syntactic Tree Pattern Analysis The analysis involves parsing essays into syntactic trees to observe pattern frequencies and patterns, focusing on AI-generated and human-written text differences. This process employs the Berkeley Neural Parser, part of the Self-Attentive Parser[5][6] suite. The code is designed to parse natural language texts, specifically our essay data, using Natural Language Processing (NLP) techniques.The function process_syntactic_tree_patterns is integral to this analysis. It utilizes
spaCy, benepar, and NLTK to dissect the syntactic structures of texts, calculating metrics like tree
depth, branching factors, nodes, leaves, and production rules. Additionally, it includes text analysis
features like token length, sentence length, and entity analysis.
Features
-
num_sentences: Counts the total number of sentences in the text, providing an overview of text segmentation. -
num_tokens: Tallies the total number of tokens (words and punctuation) in the text, reflecting the overall length. -
num_unique_lemmas: Counts distinct base forms of words (lemmas), indicating the diversity of vocabulary used. -
average_token_length: Calculates the average length of tokens, shedding light on word complexity and usage. -
average_sentence_length: Determines the average number of tokens per sentence, indicating sentence complexity. -
num_entities: Counts named entities (like people, places, organizations) recognized in the text, useful for understanding the focus and context. -
num_noun_chunks: Tallies noun phrases, providing insights into the structure and complexity of nominal groups. -
num_pos_tags: Counts the variety of parts of speech tags, reflecting grammatical diversity. -
num_distinct_entities: Determines the number of unique named entities, indicative of the text's contextual richness. -
average_entity_length: Calculates the average length of recognized entities, contributing to understanding the detail level of named references. -
average_noun_chunk_length: Measures the average length of noun chunks, indicating the complexity and composition of noun phrases. -
max_depth: Determines the maximum depth of syntactic trees in the text, a measure of syntactic complexity. -
avg_branching_factor: Calculates the average branching factor of syntactic trees, reflecting the structural complexity and diversity. -
total_nodes: Counts the total number of nodes in all syntactic trees, indicating the overall structural richness of the text. -
total_leaves: Tallies the leaves in syntactic trees, correlated with sentence simplicity or complexity. -
unique_rules: Counts the unique syntactic production rules found across all trees, indicative of syntactic variety. -
tree_complexity: Measures the complexity of the syntactic trees by comparing the number of nodes to leaves. -
depth_variability: Calculates the standard deviation of tree depths, indicating the variability in syntactic complexity across sentences.
These features collectively provide a comprehensive linguistic and structural analysis of the text, offering valuable insights into the syntactic and semantic characteristics of the processed essays.
import spacy
import benepar
import numpy as np
import pandas as pd
import logging
from collections import Counter
from nltk import Tree
from transformers import T5TokenizerFast
from tqdm import tqdm
tqdm.pandas()
import time
# Start time
#start_time = time.time()
# Configure logging
logging.basicConfig(level=logging.INFO, format='%(asctime)s - %(levelname)s - %(message)s')
logger = logging.getLogger(__name__)
import traceback
def process_syntactic_tree_patterns(df_essays):
start_time = time.time()
"""
Process a DataFrame containing essays to extract various syntactic tree pattern features.
The function uses spaCy, benepar, and NLTK to analyze syntactic structures of text,
calculating various metrics such as tree depth, branching factors, nodes, leaves,
and production rules. It also includes text analysis features like token length,
sentence length, and entity analysis.
Args:
df_essays (pandas.DataFrame): DataFrame containing a 'text' column with essays.
Returns:
pandas.DataFrame: DataFrame with additional columns for each extracted syntactic and textual feature.
"""
tokenizer = T5TokenizerFast.from_pretrained('t5-base', model_max_length=512, validate_args=False)
try:
nlp = spacy.load('en_core_web_lg') # Gotta use en_core_web_lg to use benepar_en3 for spacy 3.0
# Just add the pipe.
# nlp.add_pipe("benepar", config={"model": "benepar_en3"})
if spacy.__version__.startswith('2'):
benepar.download('benepar_en3')
nlp.add_pipe(benepar.BeneparComponent("benepar_en3"))
else:
nlp.add_pipe("benepar", config={"model": "benepar_en3"})
except Exception as e:
logger.error(f"Failed to load spaCy model: {e}")
return df_essays
def spacy_to_nltk_tree(node):
if node.n_lefts + node.n_rights > 0:
return Tree(node.orth_, [spacy_to_nltk_tree(child) for child in node.children])
else:
return node.orth_
def tree_depth(node):
if not isinstance(node, Tree):
return 0
else:
return 1 + max(tree_depth(child) for child in node)
def tree_branching_factor(node):
if not isinstance(node, Tree):
return 0
else:
return len(node)
def count_nodes(node):
if not isinstance(node, Tree):
return 1
else:
return 1 + sum(count_nodes(child) for child in node)
def count_leaves(node):
if not isinstance(node, Tree):
return 1
else:
return sum(count_leaves(child) for child in node)
def production_rules(node):
rules = []
if isinstance(node, Tree):
rules.append(node.label())
for child in node:
rules.extend(production_rules(child))
return rules
def count_labels_in_tree(tree, label):
if not isinstance(tree, Tree):
return 0
count = 1 if tree.label() == label else 0
for subtree in tree:
count += count_labels_in_tree(subtree, label)
return count
def count_phrases_by_label(trees, label, doc):
if label == 'NP':
noun_phrases = [chunk.text for chunk in doc.noun_chunks]
return noun_phrases
else:
return sum(count_labels_in_tree(tree, label) for tree in trees if isinstance(tree, Tree))
def count_subtrees_by_label(trees, label):
return sum(count_labels_in_tree(tree, label) for tree in trees if isinstance(tree, Tree))
def average_phrase_length(trees):
lengths = [len(tree.leaves()) for tree in trees if isinstance(tree, Tree)]
return np.mean(lengths) if lengths else 0
def subtree_height(tree, side):
if not isinstance(tree, Tree) or not tree:
return 0
if side == 'left':
return 1 + subtree_height(tree[0], side)
elif side == 'right':
return 1 + subtree_height(tree[-1], side)
def average_subtree_height(trees):
heights = [tree_depth(tree) for tree in trees if isinstance(tree, Tree)]
return np.mean(heights) if heights else 0
def pos_tag_distribution(trees):
pos_tags = [tag for tree in trees for word, tag in tree.pos()]
return Counter(pos_tags)
def process_tree_or_string(obj):
if isinstance(obj, Tree):
return obj.height()
else:
return None
def syntactic_ngrams(tree):
ngrams = []
if isinstance(tree, Tree):
ngrams.extend(list(nltk.ngrams(tree.pos(), 2)))
return ngrams
for index, row in df_essays.iterrows():
text = row['text']
try:
doc = nlp(text)
trees = [spacy_to_nltk_tree(sent.root) for sent in doc.sents if len(tokenizer.tokenize(sent.text)) < 512]
trees = [tree for tree in trees if isinstance(tree, Tree)]
# Extract features
depths = [tree_depth(tree) for tree in trees if isinstance(tree, Tree)]
branching_factors = [tree_branching_factor(tree) for tree in trees if isinstance(tree, Tree)]
nodes = [count_nodes(tree) for tree in trees if isinstance(tree, Tree)]
leaves = [count_leaves(tree) for tree in trees if isinstance(tree, Tree)]
rules = [production_rules(tree) for tree in trees if isinstance(tree, Tree)]
rule_counts = Counter([rule for sublist in rules for rule in sublist])
# Text analysis features
num_sentences = len(list(doc.sents))
num_tokens = len(doc)
unique_lemmas = set([token.lemma_ for token in doc])
total_token_length = sum(len(token.text) for token in doc)
average_token_length = total_token_length / num_tokens if num_tokens > 0 else 0
average_sentence_length = num_tokens / num_sentences if num_sentences > 0 else 0
num_entities = len(doc.ents)
num_noun_chunks = len(list(doc.noun_chunks))
pos_tags = [token.pos_ for token in doc]
num_pos_tags = len(set(pos_tags))
distinct_entities = set([ent.text for ent in doc.ents])
total_entity_length = sum(len(ent.text) for ent in doc.ents)
average_entity_length = total_entity_length / num_entities if num_entities > 0 else 0
total_noun_chunk_length = sum(len(chunk.text) for chunk in doc.noun_chunks)
average_noun_chunk_length = total_noun_chunk_length / num_noun_chunks if num_noun_chunks > 0 else 0
ngrams = []
for tree in trees:
ngrams.extend(syntactic_ngrams(tree))
# Assign calculated feature values to the DataFrame
df_essays.at[index, 'num_sentences'] = num_sentences
df_essays.at[index, 'num_tokens'] = num_tokens
df_essays.at[index, 'num_unique_lemmas'] = len(unique_lemmas)
df_essays.at[index, 'average_token_length'] = average_token_length
df_essays.at[index, 'average_sentence_length'] = average_sentence_length
df_essays.at[index, 'num_entities'] = num_entities
df_essays.at[index, 'num_noun_chunks'] = num_noun_chunks
df_essays.at[index, 'num_pos_tags'] = num_pos_tags
df_essays.at[index, 'num_distinct_entities'] = len(distinct_entities)
df_essays.at[index, 'average_entity_length'] = average_entity_length
df_essays.at[index, 'average_noun_chunk_length'] = average_noun_chunk_length
df_essays.at[index, 'max_depth'] = max(depths) if depths else 0
df_essays.at[index, 'avg_branching_factor'] = np.mean(branching_factors) if branching_factors else 0
df_essays.at[index, 'total_nodes'] = sum(nodes)
df_essays.at[index, 'total_leaves'] = sum(leaves)
df_essays.at[index, 'unique_rules'] = len(rule_counts)
df_essays.at[index, 'most_common_rule'] = rule_counts.most_common(1)[0][0] if rule_counts else None
df_essays.at[index, 'tree_complexity'] = sum(nodes) / sum(leaves) if leaves else 0
df_essays.at[index, 'depth_variability'] = np.std(depths)
#df_essays.at[index, 'subtree_freq_dist'] = Counter([' '.join(node.leaves()) for tree in trees for node in tree.subtrees() if isinstance(node, Tree)])
df_essays.at[index, 'tree_height_variability'] = np.std([subtree_height(tree, 'left') for tree in trees if isinstance(tree, Tree)])
#df_essays.at[index, 'pos_tag_dist'] = pos_tag_distribution(trees)
#df_essays.at[index, 'syntactic_ngrams'] = ngrams
except Exception as e:
logger.error(f"Error processing text: {e}")
traceback.print_exc()
# Assign NaNs in case of error
# df_essays.at[index, 'num_sentences'] = np.nan
# ... Assign NaNs for other features ...
return df_essays
#%timeit
# Usage
try:
print("Step 7: process_syntactic_tree_patterns")
df_essays = process_syntactic_tree_patterns(df_essays)
end_time = time.time()
# Calculate duration
duration = end_time - start_time
except Exception as e:
logger.error(f"ERROR: process_syntactic_tree_patterns: {e}")
# Explain the code in the cell. Add this line to each cell
if(CFG.EXPLAIN_CODE):
explain_code(_i)
Step 7: process_syntactic_tree_patterns
You're using a T5TokenizerFast tokenizer. Please note that with a fast tokenizer, using the `__call__` method is faster than using a method to encode the text followed by a call to the `pad` method to get a padded encoding.
/Users/lange/anaconda3/envs/py39/lib/python3.9/site-packages/torch/distributions/distribution.py:53: UserWarning:
<class 'torch_struct.distributions.TreeCRF'> does not define `arg_constraints`. Please set `arg_constraints = {}` or initialize the distribution with `validate_args=False` to turn off validation.
/Users/lange/anaconda3/envs/py39/lib/python3.9/site-packages/torch/distributions/distribution.py:53: UserWarning:
<class 'torch_struct.distributions.TreeCRF'> does not define `arg_constraints`. Please set `arg_constraints = {}` or initialize the distribution with `validate_args=False` to turn off validation.
categories_to_plot = ['num_sentences','num_tokens','num_unique_lemmas','average_token_length','average_sentence_length','num_entities','num_noun_chunks','num_pos_tags','num_distinct_entities','average_entity_length','average_noun_chunk_length','max_depth','avg_branching_factor','total_nodes','total_leaves','unique_rules','tree_complexity','depth_variability']
plot_feature_distribution(df_essays, categories_to_plot,True)
def sanity_check():
columns_with_nan = df_essays.columns[df_essays.isna().any()].tolist()
nan_count = df_essays[columns_with_nan].isna().sum()
print(nan_count)
for column, count in zip(columns_with_nan, nan_count):
print(f"Column '{column}' has {count} NaN value(s).")
assert nan_count.sum() == 0, "NaN values found in the DataFrame."
print("There are no missing values in df_essays.")
df_essays.dropna(inplace=True)
sanity_check()
Series([], dtype: float64)
There are no missing values in df_essays.
# Create a deep copy so i can use the original df_essays later
df_essays_copy = df_essays.copy(deep=True) ## for now
from sklearn.preprocessing import MinMaxScaler, StandardScaler
def scale_columns(df, columns_to_scale, scaler=None, scale_type='MinMaxScaler'):
"""
Scale the specified columns in a DataFrame and add a suffix to the column names.
Args:
df (pandas.DataFrame): The DataFrame to scale.
columns_to_scale (list): List of column names to scale.
scaler (object, optional): Scaler object to use for scaling. If None, a new scaler object will be created.
scale_type (str, optional): The type of scaler to use. Default is 'MinMaxScaler'. Options: 'MinMaxScaler', 'StandardScaler'.
Returns:
pandas.DataFrame: The full DataFrame with scaled columns added.
pandas.DataFrame: A separate DataFrame with only the specified columns scaled.
object: The scaler object used for scaling.
"""
if scale_type == 'MinMaxScaler':
scaler = MinMaxScaler() if scaler is None else scaler
elif scale_type == 'StandardScaler':
scaler = StandardScaler() if scaler is None else scaler
else:
raise ValueError("Invalid scale_type. Options: 'MinMaxScaler', 'StandardScaler'")
scaled_columns = scaler.fit_transform(df[columns_to_scale])
scaled_df = pd.DataFrame(scaled_columns, columns=[col + '_scaled' for col in columns_to_scale])
full_df = pd.concat([df.drop(columns=columns_to_scale), scaled_df], axis=1)
return full_df, scaled_df, scaler
import joblib
columns_to_scale = ['flesch_kincaid_grade', 'gunning_fog', 'coleman_liau_index', 'smog_index', 'ari', 'dale_chall', 'textual_entropy', 'semantic_density', 'semantic_flow_variability']
readability_scaled_backin_df, readability_scaled_df, readability_scaler = scale_columns(df_essays_copy, columns_to_scale, scale_type='MinMaxScaler')
joblib.dump(readability_scaler, f'{CFG.SCRATCH_PATH}/scaler_semantic_features.pkl', compress=True)
['scratch/scaler_semantic_features.pkl']
columns_to_scale = ['help','office','dance','money','wedding','domestic_work','sleep','medical_emergency','cold','hate','cheerfulness','aggression','occupation','envy','anticipation','family','vacation','crime','attractive','masculine','prison','health','pride','dispute','nervousness','government','weakness','horror','swearing_terms','leisure','suffering','royalty','wealthy','tourism','furniture','school','magic','beach','journalism','morning','banking','social_media','exercise','night','kill','blue_collar_job','art','ridicule','play','computer','college','optimism','stealing','real_estate','home','divine','sexual','fear','irritability','superhero','business','driving','pet','childish','cooking','exasperation','religion','hipster','internet','surprise','reading','worship','leader','independence','movement','body','noise','eating','medieval','zest','confusion','water','sports','death','healing','legend','heroic','celebration','restaurant','violence','programming','dominant_heirarchical','military','neglect','swimming','exotic','love','hiking','communication','hearing','order','sympathy','hygiene','weather','anonymity','trust','ancient','deception','fabric','air_travel','fight','dominant_personality','music','vehicle','politeness','toy','farming','meeting','war','speaking','listen','urban','shopping','disgust','fire','tool','phone','gain','sound','injury','sailing','rage','science','work','appearance','valuable','warmth','youth','sadness','fun','emotional','joy','affection','traveling','fashion','ugliness','lust','shame','torment','economics','anger','politics','ship','clothing','car','strength','technology','breaking','shape_and_size','power','white_collar_job','animal','party','terrorism','smell','disappointment','poor','plant','pain','beauty','timidity','philosophy','negotiate','negative_emotion','cleaning','messaging','competing','law','friends','payment','achievement','alcohol','liquid','feminine','weapon','children','monster','ocean','giving','contentment','writing','rural','positive_emotion','musical']
psycho_scaled_df_backin_df, psycho_scaled_df, psycho_scaler = scale_columns(df_essays_copy, columns_to_scale, scale_type='MinMaxScaler')
joblib.dump(psycho_scaler, f'{CFG.SCRATCH_PATH}/scaler_psycho_features.pkl', compress=True)
['scratch/scaler_psycho_features.pkl']
columns_to_scale = ['num_sentences', 'num_tokens', 'num_unique_lemmas', 'average_token_length', 'average_sentence_length', 'num_entities', 'num_noun_chunks', 'num_pos_tags', 'num_distinct_entities', 'average_entity_length', 'average_noun_chunk_length', 'max_depth', 'avg_branching_factor', 'total_nodes', 'total_leaves', 'unique_rules', 'tree_complexity', 'depth_variability']
tree_feature_scaler_backin_df, tree_features_scaled_df, tree_feature_scaler = scale_columns(df_essays_copy, columns_to_scale, scale_type='MinMaxScaler')
joblib.dump(tree_feature_scaler, f'{CFG.SCRATCH_PATH}/scaler_tree_features.pkl', compress=True)
['scratch/scaler_tree_features.pkl']
final_features_df = pd.concat([readability_scaled_df,tree_features_scaled_df,psycho_scaled_df], axis=1)
print("Shape df_essays_copy: " + str(df_essays_copy.shape))
print("Semantic Features Scaled: " + str(final_features_df.shape))
#final_features_df.head()
Shape df_essays_copy: (3999, 227)
Semantic Features Scaled: (3999, 221)
if(CFG.CLEARML_ON):
# These are the final before uploading and starting the modeling process.
upload_dataset_from_dataframe(final_features_df,"training_with_features_scaled",
'LLM-detect-ai-gen-text-LIVE/dev/notebook/preprocess',
"Training Data with Features, Post the scaling",
["training_with_features_scaled","feature"],
"scratch/training_with_features_scaled.pkl")
upload_dataset_from_dataframe(df_essays_copy,"training_with_features",
'LLM-detect-ai-gen-text-LIVE/dev/notebook/preprocess',
"Training Data with Features, Before Scaling",
["training_with_features","feature"],
"scratch/training_with_features.pkl")
clearml_handler.task.get_logger().report_table(title='training_with_features_scaled',series='Train Essays Features',
iteration=0,table_plot=final_features_df)
clearml_handler.task.close()
Model Overview
-
BertForSequenceClassification:
- Architecture: BERT (Bidirectional Encoder Representations from Transformers) for sequence classification. [4]
- Process: Involves data pre-processing, data loading, and training with hyperparameter optimization.
- Metrics: Trained and monitored using accuracy, precision, recall, F1 score, and AUC.
-
BertModel + BiLSTM:
- Architecture: The model is composed of the BertModel layer followed by BiLSTM layers. This is further connected to a dropout layer for regularization, a fully connected linear layer with ReLU activation, and a final linear layer for classification.
-
Explainable Boosting Machine (EBM) for Feature Classification:
- Type: Glass-box model, notable for interpretability and effectiveness.
- Function: Classifies based on extracted features from the essays.
- Configuration: Includes settings for interaction depth, learning rate, and validation size.
- Insights: Provides understanding of feature importance and model behavior.
- Causality This is a 'casual' model and the EBM is for helping dertermine causality aloong with our feature stats
Ensemble Approach
- Final Output Calculation: The outputs of each model are summed and then averaged to determine the ensemble's final output.
def sanity_check_2():
# Check the number of missing values
assert final_features_df.isnull().sum().sum() == 0, "There are missing values in final_features_df."
print("There are no missing values in final_features_df.")
# Check the data types
assert final_features_df.dtypes.unique().tolist() == [np.float64], "The data types of final_features_df are incorrect."
print("Data types of final_features_df are correct.")
# compare row count and assert on error
assert df_essays_copy.shape[0] == final_features_df.shape[0], "Row count mismatch between df_essays_copy and final_features_df"
print("Row count between df_essays_copy and final_features_df is correct")
# Check the range of values
# assert final_features_df.max().max() <= 1 and final_features_df.min().min() >= 0, "The values in final_features_df are not between 0 and 1."
# print("All values in final_features_df are between 0 and 1.")
sanity_check_2() # I had to take a week off after sanity_check_1() :)
There are no missing values in final_features_df.
Data types of final_features_df are correct.
Row count between df_essays_copy and final_features_df is correct
🤗 BERT for Sequence Classification
bert-base-uncased
Leveraging the 🤗 bert-base-uncased model, this section integrates the BertForSequenceClassification for efficient text sequence classification, combining BERT's advanced language processing with a streamlined, single-layer architecture for optimal simplicity and efficacy
Overview
- Model Type: BertForSequenceClassification, adapted from the pre-trained BERT model.
- Architecture: Integrates BERT's transformer layers with a single linear layer for classification.
- Design Advantage: Leverages BERT's advanced language understanding while ensuring efficiency in classification tasks.
Rationale
- Pre-trained Foundation: Utilizes the foundational BERT model pre-trained on a vast corpus. This approach leverages existing rich text representations, reducing the need for extensive training data and time.
- Simplicity and Efficiency Balance: Achieves a balance between simplicity and operational efficiency. The single linear layer addition to BERT's framework allows for effective handling of various classification tasks without overly complicating the model or extending training duration.
import torch
import pandas as pd
import numpy as np
from torch.utils.data import DataLoader
from transformers import BertTokenizer, BertForSequenceClassification, TrainingArguments
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score, roc_auc_score, confusion_matrix
import optuna
import logging
from torch.utils.tensorboard import SummaryWriter
import time
import random
# need to reconfig to error. with logging.INFO it's madness
logging.basicConfig(level=logging.ERROR)
logger = logging.getLogger(__name__)
model_path = CFG.BERT_MODEL
class TextDataset(torch.utils.data.IterableDataset):
def __init__(self, dataframe, tokenizer, max_length):
self.dataframe = dataframe
self.tokenizer = tokenizer
self.max_length = max_length
def __iter__(self):
for index, row in self.dataframe.iterrows():
text = row['text']
label = row['label']
# Encoding the text - BERT style!
encoding = self.tokenizer.encode_plus(
text,
add_special_tokens=True,
max_length=self.max_length,
padding='max_length',
truncation=True,
return_attention_mask=True,
return_tensors='pt'
)
input_ids = encoding['input_ids'].view(-1)
attention_mask = encoding['attention_mask'].view(-1)
yield input_ids, attention_mask, torch.tensor(label)
def __len__(self):
return len(self.dataframe)
class BertClassifier:
def __init__(self, training_args, model_config):
self.training_args = training_args
self.model_config = model_config
self.df = df_essays_copy
self.tokenizer = BertTokenizer.from_pretrained(CFG.BERT_MODEL, do_lower_case=model_config['do_lower_case'])
self.model = BertForSequenceClassification.from_pretrained(
CFG.BERT_MODEL,
num_labels=model_config['num_labels'],
output_attentions=model_config['output_attentions'],
output_hidden_states=model_config['output_hidden_states'],
)
# Freeze the pre-trained layers -- only update weigts for BertForSequenceClassification
# This destroys the model. We'll figure out why later, but for now -- gradient.
# for param in self.model.bert.parameters():
# param.requires_grad = False
self.run_name = f"run_{int(time.time())}"
self.writer = SummaryWriter(log_dir=f'{CFG.SCRATCH_PATH}/logs/bert_sequence_classifier/{self.run_name}')
self.model.to(training_args.device)
self.optimizer = torch.optim.AdamW(self.model.parameters(), lr=training_args.learning_rate, eps=training_args.adam_epsilon)
def _prepare_data(self):
test_ratio, val_ratio = 0.2, 0.2
train_val_idx, test_idx = train_test_split(np.arange(len(self.df)), test_size=test_ratio, shuffle=True, stratify=self.df['label'])
train_idx, val_idx = train_test_split(train_val_idx, test_size=val_ratio / (1 - test_ratio), shuffle=True, stratify=self.df.iloc[train_val_idx]['label'])
self.train_dataset = TextDataset(self.df.iloc[train_idx], self.tokenizer, self.model_config['max_length'])
self.val_dataset = TextDataset(self.df.iloc[val_idx], self.tokenizer, self.model_config['max_length'])
self.test_dataset = TextDataset(self.df.iloc[test_idx], self.tokenizer, self.model_config['max_length'])
self.train_dataloader = DataLoader(self.train_dataset, batch_size=self.training_args.per_device_train_batch_size)
self.validation_dataloader = DataLoader(self.val_dataset, batch_size=self.training_args.per_device_eval_batch_size)
self.test_dataloader = DataLoader(self.test_dataset, batch_size=self.training_args.per_device_eval_batch_size)
def train(self,trial):
self.model.train()
for epoch in tqdm(range(self.training_args.num_train_epochs), desc='Epoch'):
tr_loss = 0
for step, batch in enumerate(self.train_dataloader):
#print(f'batch size_{len(batch)}')
b_input_ids, b_input_mask, b_labels = batch
b_input_ids, b_input_mask, b_labels = b_input_ids.to(self.training_args.device), b_input_mask.to(self.training_args.device), b_labels.to(self.training_args.device)
self.optimizer.zero_grad()
outputs = self.model(b_input_ids, token_type_ids=None, attention_mask=b_input_mask, labels=b_labels)
train_loss = outputs.loss
train_loss.backward()
self.optimizer.step()
tr_loss += train_loss.item()
# Add training step loss to TensorBoard with the run name
#self.writer.add_scalar(f'{self.run_name}/Training Step Loss', tr_loss / len(self.train_dataloader), step)
accuracy, precision, recall, f1, auc, report = self.evaluate(phase='Validation', epoch=epoch)
#validation metrics to TensorBoard with the run name
self.writer.add_scalar(f'{self.run_name}/Validation Accuracy', accuracy, epoch)
self.writer.add_scalar(f'{self.run_name}/Validation Precision', precision, epoch)
self.writer.add_scalar(f'{self.run_name}/Validation Recall', recall, epoch)
self.writer.add_scalar(f'{self.run_name}/Validation F1', f1, epoch)
self.writer.add_scalar(f'{self.run_name}/Validation AUC', auc, epoch)
print(f"Validation Accuracy: {accuracy:.4f}")
print(f"precision: {precision:.4f}")
print(f"recall: {recall:.4f}")
print(f"F1: {f1:.4f}")
print(f"auc: {auc:.4f}")
print(report)
print(f"Epoch {epoch} - Train loss: {tr_loss / len(self.train_dataloader)}")
if(CFG.RETRAIN == False):
torch.save(self.model.state_dict(), f"{CFG.SCRATCH_PATH}/bert_sequence_classifier_{trial.number}_epoch_{training_args.num_train_epochs - 1}.pt")
def test(self, phase='Test', epoch=0):
if self.test_dataloader is None:
raise ValueError("Test DataLoader not set. Please set up a test DataLoader before calling test().")
self.model.eval()
all_predictions, all_true_labels = [], []
for batch in self.test_dataloader:
b_input_ids, b_input_mask, b_labels = batch
b_input_ids, b_input_mask, b_labels = b_input_ids.to(self.training_args.device), b_input_mask.to(self.training_args.device), b_labels.to(self.training_args.device)
with torch.no_grad():
outputs = self.model(b_input_ids, token_type_ids=None, attention_mask=b_input_mask)
logits = outputs.logits
predictions = torch.argmax(logits, dim=1).cpu().numpy()
labels = b_labels.cpu().numpy()
all_predictions.extend(predictions)
all_true_labels.extend(labels)
accuracy = accuracy_score(all_true_labels, all_predictions)
precision = precision_score(all_true_labels, all_predictions, average='binary')
recall = recall_score(all_true_labels, all_predictions, average='binary')
f1 = f1_score(all_true_labels, all_predictions, average='binary')
auc = roc_auc_score(all_true_labels, all_predictions)
confusion = confusion_matrix(all_true_labels, all_predictions)
# Add metrics to TensorBoard with the run name
self.writer.add_scalar(f'{self.run_name}/{phase} Test Accuracy', accuracy, epoch)
self.writer.add_scalar(f'{self.run_name}/{phase} Test Precision', precision, epoch)
self.writer.add_scalar(f'{self.run_name}/{phase} Test Recall', recall, epoch)
self.writer.add_scalar(f'{self.run_name}/{phase} Test F1', f1, epoch)
self.writer.add_scalar(f'{self.run_name}/{phase} Test AUC', auc, epoch)
logger.info(f"Epoch {epoch} - {phase} Test Metrics - Accuracy: {accuracy}, Precision: {precision}, Recall: {recall}, F1: {f1}")
return accuracy, precision, recall, f1, auc, confusion
def evaluate(self, phase='Validation', epoch=0):
self.model.eval()
predictions = []
actual_labels = []
with torch.no_grad():
for batch in self.validation_dataloader:
b_input_ids, b_input_mask, b_labels = batch
b_input_ids, b_input_mask, b_labels = b_input_ids.to(self.training_args.device), b_input_mask.to(self.training_args.device), b_labels.to(self.training_args.device)
with torch.no_grad():
outputs = self.model(b_input_ids, token_type_ids=None, attention_mask=b_input_mask)
logits = outputs.logits
batch_predictions = torch.argmax(logits, dim=1).cpu().numpy()
labels = b_labels.cpu().numpy()
predictions.extend(batch_predictions)
actual_labels.extend(labels)
# Calculate metrics
accuracy = accuracy_score(actual_labels, predictions)
precision = precision_score(actual_labels, predictions, average='binary', zero_division=1)
recall = recall_score(actual_labels, predictions, average='binary')
f1 = f1_score(actual_labels, predictions, average='binary', zero_division=1)
auc = roc_auc_score(actual_labels, predictions)
confusion = confusion_matrix(actual_labels, predictions)
logger.info(f"Epoch {epoch} - {phase} Metrics - Accuracy: {accuracy}, Precision: {precision}, Recall: {recall}, F1: {f1}")
return accuracy, precision, recall, f1, auc, confusion
def optimize_hyperparams(df, model_config, training_args, n_trials=10):
def objective(trial):
# Define hyperparameters to tune
learning_rate = trial.suggest_float("learning_rate", 1e-5, 5e-5)
batch_size = trial.suggest_categorical("batch_size", [8, 16, 32])
#num_train_epochs = trial.suggest_int("num_train_epochs", 2, 5)
local_training_args = training_args
local_training_args.learning_rate = learning_rate
local_training_args.per_device_train_batch_size = batch_size
local_training_args.num_train_epochs = training_args.num_train_epochs
classifier = BertClassifier(local_training_args, model_config)
classifier._prepare_data()
# %reload_ext tensorboard
# %tensorboard --logdir=./logs/bert_sequence_classifier/
#%%timeit
classifier.train(trial)
# Evaluate the modell
accuracy, precision, recall, f1, auc, confusion = classifier.test()
print("Test Metrics with Best Hyperparameters:")
print(f"AUC: {auc}, Precision: {precision}, Recall: {recall}, F1 Score: {f1}")
print("Confusion Matrix:")
print(confusion)
return auc
study = optuna.create_study(direction="maximize")
study.optimize(objective, n_trials=n_trials)
print("Best trial:")
trial = study.best_trial
print(f" Value: {trial.value}")
print(" Params: ")
for key, value in trial.params.items():
print(f" {key}: {value}")
return study.best_trial.params
def retrain_with_best_hyperparams(df, model_config, best_params):
training_args.learning_rate = best_params["learning_rate"]
training_args.per_device_train_batch_size = best_params["batch_size"]
classifier = BertClassifier(training_args, model_config)
classifier._prepare_data()
CFG.RETRAIN = True
classifier.train(None) # Pass None as trial is not needed here
tokenizer = BertTokenizer.from_pretrained(model_config['bert_model'], do_lower_case=model_config['do_lower_case'])
# Pickle the tokenizer, study, and best model
with open(f'{CFG.SCRATCH_PATH}/bert_seq_class_tokeniser.pkl', 'wb') as f:
pickle.dump(tokenizer, f)
with open(f'{CFG.SCRATCH_PATH}/bert_seq_class_study.pkl', 'wb') as f:
pickle.dump(best_params, f)
# Save the best model
torch.save(classifier.model.state_dict(), f"{CFG.SCRATCH_PATH}/bert_sequence_classifier_best.pt")
# Main Execution
if CFG.EBM_ONLY == False:
# Configuration and Usage
model_config = {
'bert_model': CFG.BERT_MODEL,
'do_lower_case': False,
'num_labels': 2,
'output_attentions': False,
'output_hidden_states': False,
'max_length': 32,
'optuna_trials': 2,
'epochs': 5,
'device': torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')
}
# FYI - device is a non-mutable proprty of TrainingArguments. so when training_args.device is get it gets properly.
training_args = TrainingArguments(
output_dir=f'{CFG.SCRATCH_PATH}/results',
num_train_epochs=model_config['epochs'],
per_device_train_batch_size=16,
per_device_eval_batch_size=32,
warmup_steps=100,
weight_decay=0.01,
logging_dir=f'{CFG.SCRATCH_PATH}/logs/bert_sequence_classifier',
learning_rate=5e-5,
adam_epsilon=1e-8,
do_train=True,
do_eval=True,
no_cuda=False,
evaluation_strategy="epoch",
save_strategy="steps",
logging_steps=500,
save_steps=500,
)
if CFG.CLEARML_ON:
# ClearML integration (if applicable)
pass
if(CFG.EBM_ONLY == False):
# Optimize hyperparameters
best_params = optimize_hyperparams(df_essays_copy, model_config, training_args, n_trials=model_config['optuna_trials'])
# Retrain model with best hyperparameters
retrain_with_best_hyperparams(df_essays_copy, model_config, best_params)
[I 2024-01-24 17:41:49,075] A new study created in memory with name: no-name-6edcff65-644b-47c4-808b-1fcb61049fbd
Some weights of BertForSequenceClassification were not initialized from the model checkpoint at bert-base-uncased and are newly initialized: ['classifier.bias', 'classifier.weight']
You should probably TRAIN this model on a down-stream task to be able to use it for predictions and inference.
Epoch: 0%| | 0/5 [00:00<?, ?it/s]INFO:__main__:Epoch 0 - Validation Metrics - Accuracy: 0.9325, Precision: 0.9435897435897436, Recall: 0.92, F1: 0.9316455696202531
Epoch: 20%|██ | 1/5 [01:06<04:26, 66.64s/it]
Validation Accuracy: 0.9325
precision: 0.9436
recall: 0.9200
F1: 0.9316
auc: 0.9325
[[378 22]
[ 32 368]]
Epoch 0 - Train loss: 0.358369643787543
INFO:__main__:Epoch 1 - Validation Metrics - Accuracy: 0.94, Precision: 0.9756756756756757, Recall: 0.9025, F1: 0.9376623376623378
Epoch: 40%|████ | 2/5 [02:08<03:11, 63.72s/it]
Validation Accuracy: 0.9400
precision: 0.9757
recall: 0.9025
F1: 0.9377
auc: 0.9400
[[391 9]
[ 39 361]]
Epoch 1 - Train loss: 0.14328922991951307
INFO:__main__:Epoch 2 - Validation Metrics - Accuracy: 0.92625, Precision: 0.8974358974358975, Recall: 0.9625, F1: 0.9288299155609168
Epoch: 60%|██████ | 3/5 [03:07<02:03, 61.82s/it]
Validation Accuracy: 0.9263
precision: 0.8974
recall: 0.9625
F1: 0.9288
auc: 0.9262
[[356 44]
[ 15 385]]
Epoch 2 - Train loss: 0.08034678868949413
INFO:__main__:Epoch 3 - Validation Metrics - Accuracy: 0.94, Precision: 0.9756756756756757, Recall: 0.9025, F1: 0.9376623376623378
Epoch: 80%|████████ | 4/5 [04:08<01:01, 61.39s/it]
Validation Accuracy: 0.9400
precision: 0.9757
recall: 0.9025
F1: 0.9377
auc: 0.9400
[[391 9]
[ 39 361]]
Epoch 3 - Train loss: 0.04097894656782349
INFO:__main__:Epoch 4 - Validation Metrics - Accuracy: 0.91, Precision: 0.9823529411764705, Recall: 0.835, F1: 0.9027027027027026
Epoch: 100%|██████████| 5/5 [05:08<00:00, 61.75s/it]
Validation Accuracy: 0.9100
precision: 0.9824
recall: 0.8350
F1: 0.9027
auc: 0.9100
[[394 6]
[ 66 334]]
Epoch 4 - Train loss: 0.023587635420262813
INFO:__main__:Epoch 0 - Test Test Metrics - Accuracy: 0.89, Precision: 0.9785276073619632, Recall: 0.7975, F1: 0.8787878787878788
[I 2024-01-24 17:47:20,934] Trial 0 finished with value: 0.8899999999999999 and parameters: {'learning_rate': 3.600434667440913e-05, 'batch_size': 32}. Best is trial 0 with value: 0.8899999999999999.
Test Metrics with Best Hyperparameters:
AUC: 0.8899999999999999, Precision: 0.9785276073619632, Recall: 0.7975, F1 Score: 0.8787878787878788
Confusion Matrix:
[[393 7]
[ 81 319]]
Some weights of BertForSequenceClassification were not initialized from the model checkpoint at bert-base-uncased and are newly initialized: ['classifier.bias', 'classifier.weight']
You should probably TRAIN this model on a down-stream task to be able to use it for predictions and inference.
Epoch: 0%| | 0/5 [00:00<?, ?it/s]INFO:__main__:Epoch 0 - Validation Metrics - Accuracy: 0.92375, Precision: 0.908433734939759, Recall: 0.9425, F1: 0.9251533742331288
Epoch: 20%|██ | 1/5 [01:32<06:09, 92.39s/it]
Validation Accuracy: 0.9237
precision: 0.9084
recall: 0.9425
F1: 0.9252
auc: 0.9237
[[362 38]
[ 23 377]]
Epoch 0 - Train loss: 0.3173389121890068
INFO:__main__:Epoch 1 - Validation Metrics - Accuracy: 0.9425, Precision: 0.9732620320855615, Recall: 0.91, F1: 0.9405684754521965
Epoch: 40%|████ | 2/5 [02:59<04:27, 89.18s/it]
Validation Accuracy: 0.9425
precision: 0.9733
recall: 0.9100
F1: 0.9406
auc: 0.9425
[[390 10]
[ 36 364]]
Epoch 1 - Train loss: 0.11091918416864549
INFO:__main__:Epoch 2 - Validation Metrics - Accuracy: 0.935, Precision: 0.9371859296482412, Recall: 0.9325, F1: 0.9348370927318297
Epoch: 60%|██████ | 3/5 [04:24<02:54, 87.49s/it]
Validation Accuracy: 0.9350
precision: 0.9372
recall: 0.9325
F1: 0.9348
auc: 0.9350
[[375 25]
[ 27 373]]
Epoch 2 - Train loss: 0.03493080863496289
INFO:__main__:Epoch 3 - Validation Metrics - Accuracy: 0.9475, Precision: 0.9408866995073891, Recall: 0.955, F1: 0.9478908188585607
Epoch: 80%|████████ | 4/5 [05:51<01:27, 87.32s/it]
Validation Accuracy: 0.9475
precision: 0.9409
recall: 0.9550
F1: 0.9479
auc: 0.9475
[[376 24]
[ 18 382]]
Epoch 3 - Train loss: 0.0410450704190104
INFO:__main__:Epoch 4 - Validation Metrics - Accuracy: 0.9325, Precision: 0.9080188679245284, Recall: 0.9625, F1: 0.9344660194174758
Epoch: 100%|██████████| 5/5 [07:19<00:00, 87.82s/it]
Validation Accuracy: 0.9325
precision: 0.9080
recall: 0.9625
F1: 0.9345
auc: 0.9325
[[361 39]
[ 15 385]]
Epoch 4 - Train loss: 0.03605509036802687
INFO:__main__:Epoch 0 - Test Test Metrics - Accuracy: 0.93625, Precision: 0.9245742092457421, Recall: 0.95, F1: 0.93711467324291
[I 2024-01-24 17:54:52,249] Trial 1 finished with value: 0.93625 and parameters: {'learning_rate': 4.2982098663601536e-05, 'batch_size': 8}. Best is trial 1 with value: 0.93625.
Test Metrics with Best Hyperparameters:
AUC: 0.93625, Precision: 0.9245742092457421, Recall: 0.95, F1 Score: 0.93711467324291
Confusion Matrix:
[[369 31]
[ 20 380]]
Best trial:
Value: 0.93625
Params:
learning_rate: 4.2982098663601536e-05
batch_size: 8
Some weights of BertForSequenceClassification were not initialized from the model checkpoint at bert-base-uncased and are newly initialized: ['classifier.bias', 'classifier.weight']
You should probably TRAIN this model on a down-stream task to be able to use it for predictions and inference.
Epoch: 0%| | 0/5 [00:00<?, ?it/s]INFO:__main__:Epoch 0 - Validation Metrics - Accuracy: 0.925, Precision: 0.956989247311828, Recall: 0.89, F1: 0.922279792746114
Epoch: 20%|██ | 1/5 [01:30<06:03, 90.76s/it]
Validation Accuracy: 0.9250
precision: 0.9570
recall: 0.8900
F1: 0.9223
auc: 0.9250
[[384 16]
[ 44 356]]
Epoch 0 - Train loss: 0.3212550792160134
INFO:__main__:Epoch 1 - Validation Metrics - Accuracy: 0.93, Precision: 0.9432989690721649, Recall: 0.915, F1: 0.9289340101522843
Epoch: 40%|████ | 2/5 [02:58<04:26, 88.98s/it]
Validation Accuracy: 0.9300
precision: 0.9433
recall: 0.9150
F1: 0.9289
auc: 0.9300
[[378 22]
[ 34 366]]
Epoch 1 - Train loss: 0.09840513726308321
INFO:__main__:Epoch 2 - Validation Metrics - Accuracy: 0.94375, Precision: 0.9539641943734015, Recall: 0.9325, F1: 0.943109987357775
Epoch: 60%|██████ | 3/5 [04:21<02:52, 86.45s/it]
Validation Accuracy: 0.9437
precision: 0.9540
recall: 0.9325
F1: 0.9431
auc: 0.9437
[[382 18]
[ 27 373]]
Epoch 2 - Train loss: 0.06413869087429096
INFO:__main__:Epoch 3 - Validation Metrics - Accuracy: 0.93375, Precision: 0.9506493506493506, Recall: 0.915, F1: 0.9324840764331209
Epoch: 80%|████████ | 4/5 [05:44<01:25, 85.03s/it]
Validation Accuracy: 0.9337
precision: 0.9506
recall: 0.9150
F1: 0.9325
auc: 0.9337
[[381 19]
[ 34 366]]
Epoch 3 - Train loss: 0.02114799525414128
INFO:__main__:Epoch 4 - Validation Metrics - Accuracy: 0.93875, Precision: 0.948849104859335, Recall: 0.9275, F1: 0.9380530973451328
Epoch: 100%|██████████| 5/5 [07:09<00:00, 85.99s/it]
Validation Accuracy: 0.9387
precision: 0.9488
recall: 0.9275
F1: 0.9381
auc: 0.9387
[[380 20]
[ 29 371]]
Epoch 4 - Train loss: 0.03044251971266931
🤗 BERT-BiLSTM Classifier Model
Rationale
- BERT's Foundational Strength: Utilizing the pre-trained BERT layers, the model leverages BERT's deep understanding of language semantics, gained from extensive training on diverse text corpora.
- Sequence and Context Awareness with BiLSTM: The addition of BiLSTM layers enables the model to capture contextual information in both forward and backward directions, making it adept at understanding the sequence and flow of the text.
- Enhanced Text Processing Capabilities: This architecture is particularly effective for complex classification tasks where understanding the context and sequence of words is crucial.
Architecture
- Composition: The model is composed of the BertModel layer followed by BiLSTM layers. This is further connected to a dropout layer for regularization, a fully connected linear layer with ReLU activation, and a final linear layer for classification.
- BiLSTM Configuration: The BiLSTM layers are configured with customizable hidden sizes and layer counts, allowing the model to be adapted to different levels of sequence complexity.
- Loss and Optimization: The model employs CrossEntropyLoss for loss computation and uses the AdamW optimizer. It focuses on optimizing metrics like accuracy and AUC, with an emphasis on balancing precision and recall.
Adaptability and Use Cases
- Versatile for Various Text Data: Given its enhanced contextual understanding, the BERT-BiLSTM Classifier is well-suited for tasks like sentiment analysis, topic classification, and other scenarios where the sequence of text plays a significant role.
- Customization and Flexibility: The adjustable parameters of the BiLSTM layers (like hidden size and number of layers) offer flexibility, making the model adaptable to a wide range of text classification challenges.
import os
import time
import numpy as np
import torch
from torch import nn
from torch.utils.data import DataLoader, Dataset
from transformers import BertTokenizer, BertModel, AdamW, get_linear_schedule_with_warmup
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, classification_report, precision_score, recall_score, f1_score, confusion_matrix, roc_auc_score
import pandas as pd
import logging
import optuna
from torch.utils.tensorboard import SummaryWriter
from tqdm import tqdm
import seaborn as sns
import matplotlib.pyplot as plt
import pickle
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model_config = {
'bert_model_name': CFG.BERT_MODEL,
'num_classes': 2,
'max_length': 128,
'batch_size': 16,
'num_epochs': 4,
'train_data': df_essays_copy,
'num_trials': 2,
}
if(CFG.CLEARML_ON):
clearml_bertmodel_custom.task.connect(model_config)
def load_data():
texts = model_config['train_data']['text'].str.lower().tolist() # Lowercase for uncased BERT
labels = model_config['train_data']['label'].tolist()
return texts, labels
class TextClassificationDataset(Dataset):
def __init__(self, texts, labels, tokenizer, max_length):
self.encodings = tokenizer(texts, add_special_tokens=True, max_length=max_length, padding='max_length', truncation=True, return_attention_mask=True, return_tensors='pt').to(device)
self.labels = torch.tensor(labels, dtype=torch.long).to(device)
def __len__(self):
return len(self.labels)
def __getitem__(self, idx):
item = {key: val[idx] for key, val in self.encodings.items()}
item['label'] = self.labels[idx]
return item
# Bert Bi-Directional LSTM
class BERTBiLSTMClassifier(nn.Module):
def __init__(self, bert_model_name, num_classes, dropout_rate=0.1, lstm_hidden_size=128, lstm_layers=2):
super(BERTBiLSTMClassifier, self).__init__()
self.bert = BertModel.from_pretrained(bert_model_name)
self.lstm = nn.LSTM(self.bert.config.hidden_size, lstm_hidden_size, lstm_layers, batch_first=True, bidirectional=True)
self.dropout = nn.Dropout(dropout_rate)
self.fc = nn.Linear(lstm_hidden_size * 2, num_classes) # *2 for bidirectional
self.relu = nn.ReLU() # ReLU activation layer
def forward(self, input_ids, attention_mask):
outputs = self.bert(input_ids=input_ids, attention_mask=attention_mask)
sequence_output = outputs.last_hidden_state
lstm_output, (h_n, c_n) = self.lstm(sequence_output)
pooled_output = torch.cat((h_n[-2,:,:], h_n[-1,:,:]), dim = 1)
x = self.dropout(pooled_output)
x = self.relu(x) # Apply ReLU activation
x = self.fc(x)
return x
if(CFG.EBM_ONLY == False):
texts, labels = load_data()
train_texts, temp_texts, train_labels, temp_labels = train_test_split(texts, labels, test_size=0.3, random_state=42)
val_texts, test_texts, val_labels, test_labels = train_test_split(temp_texts, temp_labels, test_size=0.5, random_state=42)
tokenizer = BertTokenizer.from_pretrained(model_config['bert_model_name'])
train_dataset = TextClassificationDataset(train_texts, train_labels, tokenizer, model_config['max_length'])
val_dataset = TextClassificationDataset(val_texts, val_labels, tokenizer, model_config['max_length'])
test_dataset = TextClassificationDataset(test_texts, test_labels, tokenizer, model_config['max_length'])
train_dataloader = DataLoader(train_dataset, batch_size=model_config['batch_size'], shuffle=True)
val_dataloader = DataLoader(val_dataset, batch_size=model_config['batch_size'])
test_dataloader = DataLoader(test_dataset, batch_size=model_config['batch_size'])
run_name = f"run_{int(time.time())}"
logging.basicConfig(level=logging.INFO)
logger = logging.getLogger(__name__)
writer = SummaryWriter(log_dir=f'{CFG.SCRATCH_PATH}/logs/bertmodel_custom/{run_name}')
def train(model, data_loader, optimizer, scheduler, device, epoch):
model.train()
total_loss = 0
leng = len(data_loader)
for batch in data_loader:
optimizer.zero_grad()
input_ids = batch['input_ids'].to(device)
attention_mask = batch['attention_mask'].to(device)
labels = batch['label'].to(device)
outputs = model(input_ids=input_ids, attention_mask=attention_mask)
loss = nn.CrossEntropyLoss()(outputs, labels)
loss.backward()
optimizer.step()
scheduler.step()
total_loss += loss.item()
avg_loss = total_loss / leng
#logger.info(f"Epoch {epoch} - Training loss: {avg_loss}")
writer.add_scalar('Training Loss', avg_loss, epoch)
def evaluate(model, data_loader, device, epoch, phase='Validation'):
model.eval()
predictions = []
actual_labels = []
_labels = []
with torch.no_grad():
for batch in data_loader:
input_ids = batch['input_ids'].to(device)
attention_mask = batch['attention_mask'].to(device)
labels = batch['label'].to(device)
outputs = model(input_ids=input_ids, attention_mask=attention_mask)
_, preds = torch.max(outputs, dim=1)
predictions.extend(preds.cpu().tolist())
actual_labels.extend(labels.cpu().tolist())
accuracy = accuracy_score(actual_labels, predictions)
precision = precision_score(actual_labels, predictions, average='binary', zero_division=1)
recall = recall_score(actual_labels, predictions, average='binary')
f1 = f1_score(actual_labels, predictions, average='binary', zero_division=1)
auc = roc_auc_score(actual_labels, predictions)
conf_matrix = confusion_matrix(actual_labels, predictions)
sns.heatmap(conf_matrix, annot=True, fmt='d')
plt.title(f'{phase} Confusion Matrix')
plt.ylabel('Actual Label')
plt.xlabel('Predicted Label')
plt.savefig(f'{phase}_confusion_matrix_epoch_{epoch}.png')
plt.close()
#logger.info(f"Epoch {epoch} - {phase} Metrics - Accuracy: {accuracy}, Precision: {precision}, Recall: {recall}, F1: {f1}")
writer.add_scalar(f'{phase} Accuracy', accuracy, epoch)
writer.add_scalar(f'{phase} Precision', precision, epoch)
writer.add_scalar(f'{phase} Recall', recall, epoch)
writer.add_scalar(f'{phase} F1 Score', f1, epoch)
return accuracy, precision, recall, f1, auc, classification_report(actual_labels, predictions)
# Optuna Hyperparameter Optimization
def objective(trial):
# Suggest hyperparameters for training
learning_rate = trial.suggest_float('learning_rate', 1e-5, 5e-5)
batch_size = trial.suggest_int('batch_size', 16, 32)
# Suggest hyperparameters for model architecture
dropout_rate = trial.suggest_float('dropout_rate', 0.01, 0.1)
fc_layer_size = trial.suggest_categorical('fc_layer_size', [32, 64])
lstm_hidden_size = trial.suggest_categorical('lstm_hidden_size', [64, 128])# =128,
lstm_layers=trial.suggest_int('lstm_layers', 2, 4)
#model = BERTBiLSTMClassifier(model_config['bert_model_name'],model_config['num_classes'],dropout_rate,lstm_hidden_size )
model = BERTBiLSTMClassifier(model_config['bert_model_name'], model_config['num_classes'], dropout_rate, fc_layer_size,lstm_layers)
model.to(device)
optimizer = torch.optim.AdamW(model.parameters(), lr=learning_rate)
#total_steps = len(train_dataloader) / model_config['num_epochs'] / model_config['batch_size']
total_steps = len(train_dataloader) * model_config['num_epochs']
scheduler = get_linear_schedule_with_warmup(optimizer, num_warmup_steps=0, num_training_steps=total_steps)
best_val_auc = 0
for epoch in tqdm(range(model_config['num_epochs']), desc='Epoch'):
train(model, train_dataloader, optimizer, scheduler, device, epoch)
accuracy, precision, recall, f1, auc, report = evaluate(model, val_dataloader, device, epoch)
print(f"Validation Accuracy: {accuracy:.4f}")
print(f"precision: {precision:.4f}")
print(f"recall: {recall:.4f}")
print(f"F1: {f1:.4f}")
print(f"auc: {auc:.4f}")
print(report)
if auc > best_val_auc:
best_val_auc = auc
best_params = {
'learning_rate': learning_rate,
'dropout_rate': dropout_rate,
'fc_layer_size': fc_layer_size
}
torch.save(model.state_dict(), f"{CFG.SCRATCH_PATH}/bert_finetune_custom_{trial.number}.pt")
torch.save(best_params, f"{CFG.SCRATCH_PATH}/best_trial_params.json")
return best_val_auc
bert_best_custom_study = optuna.create_study(direction='maximize', study_name='bert_best_custom_study')
bert_best_custom_study.optimize(objective, n_trials=model_config['num_trials'])
best_trial = bert_best_custom_study.best_trial
#Load the model with the best trial
best_trial_params = bert_best_custom_study.best_trial.params
learning_rate = best_trial_params["learning_rate"]
dropout_rate = best_trial_params["dropout_rate"]
fc_layer_size = best_trial_params["fc_layer_size"]
lstm_hidden_size = best_trial_params["lstm_hidden_size"]
lstm_layers = best_trial_params["lstm_layers"]
# Pickle the tokenizer, study, and best model
with open(f'{CFG.SCRATCH_PATH}/custom_bert_tokenizer.pkl', 'wb') as f:
pickle.dump(tokenizer, f)
with open(f'{CFG.SCRATCH_PATH}/best_custom_model_study.pkl', 'wb') as f:
pickle.dump(bert_best_custom_study, f)
#Initialize the best model with the optimal hyperparameters
best_model = BERTBiLSTMClassifier(model_config['bert_model_name'], model_config['num_classes'], dropout_rate, fc_layer_size,lstm_layers)
best_model.to(device)
#Set up optimizer and scheduler for the best model
optimizer = torch.optim.AdamW(best_model.parameters(), lr=learning_rate)
total_steps = len(train_dataloader) * model_config['num_epochs']
scheduler = get_linear_schedule_with_warmup(optimizer, num_warmup_steps=0, num_training_steps=total_steps)
#Retrain the model with the best hyperparameters
for epoch in tqdm(range(model_config['num_epochs']), desc='Epoch'):
train(best_model, train_dataloader, optimizer, scheduler, device, epoch)
evaluate(best_model, val_dataloader, device, epoch)
#Save the retrained best model
torch.save(best_model.state_dict(), f"{CFG.SCRATCH_PATH}/bert_finetune_custom_best.pt")
#Print best trial details
print("Best trial:")
print(f" Value: {best_trial.value:.4f}")
print(" Params: ")
for key, value in best_trial.params.items():
print(f" {key}: {value}")
[I 2024-01-24 18:02:56,813] A new study created in memory with name: bert_best_custom_study
Epoch: 0%| | 0/4 [00:00<?, ?it/s]
Validation Accuracy: 0.8767
precision: 0.8005
recall: 1.0000
F1: 0.8892
auc: 0.8779
precision recall f1-score support
0 1.00 0.76 0.86 303
1 0.80 1.00 0.89 297
accuracy 0.88 600
macro avg 0.90 0.88 0.88 600
weighted avg 0.90 0.88 0.87 600
Epoch: 25%|██▌ | 1/4 [06:23<19:11, 383.91s/it]
Validation Accuracy: 0.9783
precision: 0.9765
recall: 0.9798
F1: 0.9782
auc: 0.9783
precision recall f1-score support
0 0.98 0.98 0.98 303
1 0.98 0.98 0.98 297
accuracy 0.98 600
macro avg 0.98 0.98 0.98 600
weighted avg 0.98 0.98 0.98 600
Epoch: 75%|███████▌ | 3/4 [18:46<06:12, 372.21s/it]
Validation Accuracy: 0.9783
precision: 0.9797
recall: 0.9764
F1: 0.9781
auc: 0.9783
precision recall f1-score support
0 0.98 0.98 0.98 303
1 0.98 0.98 0.98 297
accuracy 0.98 600
macro avg 0.98 0.98 0.98 600
weighted avg 0.98 0.98 0.98 600
Epoch: 100%|██████████| 4/4 [24:44<00:00, 371.22s/it]
[I 2024-01-24 18:27:42,895] Trial 0 finished with value: 0.9783478347834783 and parameters: {'learning_rate': 4.305004023823152e-05, 'batch_size': 25, 'dropout_rate': 0.0456841451290286, 'fc_layer_size': 32, 'lstm_hidden_size': 64, 'lstm_layers': 3}. Best is trial 0 with value: 0.9783478347834783.
Validation Accuracy: 0.9700
precision: 0.9515
recall: 0.9899
F1: 0.9703
auc: 0.9702
precision recall f1-score support
0 0.99 0.95 0.97 303
1 0.95 0.99 0.97 297
accuracy 0.97 600
macro avg 0.97 0.97 0.97 600
weighted avg 0.97 0.97 0.97 600
Epoch: 0%| | 0/4 [00:00<?, ?it/s]
Validation Accuracy: 0.9617
precision: 0.9724
recall: 0.9495
F1: 0.9608
auc: 0.9615
precision recall f1-score support
0 0.95 0.97 0.96 303
1 0.97 0.95 0.96 297
accuracy 0.96 600
macro avg 0.96 0.96 0.96 600
weighted avg 0.96 0.96 0.96 600
Epoch: 50%|█████ | 2/4 [11:34<11:30, 345.01s/it]
Validation Accuracy: 0.9533
precision: 0.9468
recall: 0.9596
F1: 0.9532
auc: 0.9534
precision recall f1-score support
0 0.96 0.95 0.95 303
1 0.95 0.96 0.95 297
accuracy 0.95 600
macro avg 0.95 0.95 0.95 600
weighted avg 0.95 0.95 0.95 600
Validation Accuracy: 0.9700
precision: 0.9635
recall: 0.9764
F1: 0.9699
auc: 0.9701
precision recall f1-score support
0 0.98 0.96 0.97 303
1 0.96 0.98 0.97 297
accuracy 0.97 600
macro avg 0.97 0.97 0.97 600
weighted avg 0.97 0.97 0.97 600
Epoch: 100%|██████████| 4/4 [23:44<00:00, 356.23s/it]
[I 2024-01-24 18:51:28,436] Trial 1 finished with value: 0.9700636730339701 and parameters: {'learning_rate': 3.6826657838303954e-05, 'batch_size': 20, 'dropout_rate': 0.06958928004555641, 'fc_layer_size': 32, 'lstm_hidden_size': 64, 'lstm_layers': 4}. Best is trial 0 with value: 0.9783478347834783.
Validation Accuracy: 0.9700
precision: 0.9697
recall: 0.9697
F1: 0.9697
auc: 0.9700
precision recall f1-score support
0 0.97 0.97 0.97 303
1 0.97 0.97 0.97 297
accuracy 0.97 600
macro avg 0.97 0.97 0.97 600
weighted avg 0.97 0.97 0.97 600
Epoch: 100%|██████████| 4/4 [22:36<00:00, 339.08s/it]
Best trial:
Value: 0.9783
Params:
learning_rate: 4.305004023823152e-05
batch_size: 25
dropout_rate: 0.0456841451290286
fc_layer_size: 32
lstm_hidden_size: 64
lstm_layers: 3
🔬 Explainable Boosting Machine (EBM)
A Balance of Predictive Power and Interpretability
EBMs function like a choir 🎶, where each data feature represents a unique voice. These features individually contribute to the overall prediction, akin to each voice adding to the choir's harmony. This additive model approach ensures that the impact of each feature is distinct and quantifiable.
Overview of EBMs
EBMs are an advanced form of Generalized Additive Models (GAMs). They enhance predictive power while maintaining high interpretability by combining traditional machine learning techniques with the additive structure of GAMs. This design allows for a clear understanding of the influence of individual features and their combinations on the predicted outcome.
Key Components of EBM
-
Formula Representation: $$ g(E[y]) = \beta_0 + \sum f_i(x_i) + \sum f_{i,j}(x_i, x_j) $$
- g(E[y]): Represents the transformed expected value of the target variable y.
- β₀: The intercept term, indicating the baseline prediction without feature consideration.
- Σ fᵢ(xᵢ): Sum of functions for individual features, showing each feature's independent effect.
- Σ fᵢⱼ(xᵢ, xⱼ): Sum of pairwise interaction terms, capturing feature interactions.
-
Training Process:
- EBMs learn shape functions fᵢ(xᵢ) and fᵢⱼ(xᵢ, xⱼ) for each feature and feature interaction through boosting. This technique incrementally refines the model by addressing previous iteration errors.
-
Interpretability:
- Post-training, the model's terms are individually examined to understand their impact on predictions. This granular approach allows for detailed insights into how each feature (and pairs of features) affects the outcome.
-
Feature Importance:
- The magnitude and shape of the learned functions provide a direct measure of feature significance. This aspect is crucial for identifying key influencers in the prediction process.
-
Flexibility and Complexity:
- Despite its interpretability, EBMs can model complex nonlinear relationships and interactions, surpassing the capabilities of traditional linear models.
☃ EBMs present a unique combination of high interpretability and predictive accuracy. This makes them ideal for scenarios where understanding the reasoning behind model decisions is as critical as the decisions themselves.
## ClearML Setup
import pickle
if(CFG.CLEARML_ON):
clearml_ebm = ClearMLTaskHandler(
project_name='LLM-detect-ai-gen-text-LIVE/dev/notebook/models/EBM',
task_name='Explainable Boosting Machine Model',
)
clearml_ebm.task.auto_connect_frameworks={
'matplotlib': True, 'tensorflow': True, 'tensorboard': True, 'pytorch': True, 'scikit': True,
'detect_repository': True, 'joblib': True,
}
import pandas as pd
import textwrap
from sklearn.model_selection import train_test_split
from interpret.perf import ROC, PR
# import openai
# import guidance
import os
from interpret.glassbox import ExplainableBoostingClassifier
from interpret import show
from plotly.offline import init_notebook_mode
init_notebook_mode(connected=True)
# import t2ebm
labels = df_essays['label'].tolist()
# Which feature Group
do_standard_scale = False
use_feature_list = False
features = final_features_df
# To see changes in the feature makeup of the model, increase the interactions. We are not. Don't want extra features we did not create, yet.
model_config = {
'feature_names': features.columns.tolist(),
'feature_types': None,
'exclude': [],
'max_bins': 255,
'validation_size': 0.20,
'outer_bags': 25, # recommended for best accuracy
'inner_bags': 25, # recommended for best accuracy
'learning_rate': 0.01,
'greediness': 0.0,
'smoothing_rounds': 0,
'early_stopping_rounds': 50,
'early_stopping_tolerance': 0.0001,
'objective': 'roc_auc',
'n_jobs': -2,
'random_state': 42
}
if(CFG.CLEARML_ON):
clearml_ebm.task.connect(model_config, name="model_config")
X_train, X_test, y_train, y_test = train_test_split(features, labels, test_size=0.2, random_state=42)
from interpret import show
from interpret.data import ClassHistogram, Marginal
hist = ClassHistogram(feature_names=features, feature_types=None).explain_data(X_train, y_train, name = 'Train Data ClassHistogram')
show(hist)
from interpret.glassbox import ExplainableBoostingClassifier
from sklearn.model_selection import RandomizedSearchCV
param_test = {'learning_rate': [0.001,0.005,0.01,0.03],
'max_rounds': [5000,10000,15000,20000],
'min_samples_leaf': [2,3,5],
'max_leaves': [3,5,10]}
n_HP_points_to_test=10
ebm_clf = ExplainableBoostingClassifier(feature_names=features.columns.tolist(), feature_types=None, n_jobs=- 2, random_state=42)
ebm_gs = RandomizedSearchCV(
estimator=ebm_clf,
param_distributions=param_test,
n_iter=n_HP_points_to_test,
scoring="roc_auc",
cv=3,
refit=True,
random_state=314,
verbose=False,
)
model = ebm_gs.fit(X_train, y_train)
# So we can use the EBM and all it's inherent functinality. Retrain with the best params.
merged_dict = {**ebm_gs.best_params_, **model_config}
ebm = ExplainableBoostingClassifier(feature_names=merged_dict["feature_names"],
feature_types=None,
interactions=0,
exclude=[],
max_bins=255,
validation_size=0.15,
outer_bags=25,
inner_bags=25,
learning_rate=merged_dict["learning_rate"],
greediness=0.0,
smoothing_rounds=0,
max_rounds=merged_dict["max_rounds"],
early_stopping_rounds=50,
early_stopping_tolerance=0.0001,
min_samples_leaf=merged_dict["min_samples_leaf"],
max_leaves=merged_dict["max_leaves"],
n_jobs=- 2,
random_state=42)
ebm.fit(X_train, y_train)
ebm.score(X_test, y_test)
ebm_perf = ROC(ebm.predict_proba).explain_perf(X_test, y_test, name='EBM')
ebm_perf_pr = PR(ebm.predict_proba).explain_perf(X_test, y_test, name='EBM Precision Recall')
show(ebm_perf)
show(ebm_perf_pr)
with open(f'{CFG.SCRATCH_PATH}/ebm.pkl', 'wb') as f:
pickle.dump(ebm, f)
from interpret import show
ebm_global = ebm.explain_global()
show(ebm_global)
/Users/lange/anaconda3/envs/py39/lib/python3.9/site-packages/interpret/visual/_udash.py:117: FutureWarning:
Series.__getitem__ treating keys as positions is deprecated. In a future version, integer keys will always be treated as labels (consistent with DataFrame behavior). To access a value by position, use `ser.iloc[pos]`
# Lets look at a couple
ebm_local = ebm.explain_local(X_test[10:15], y_test[10:15])
show(ebm_local)
/Users/lange/anaconda3/envs/py39/lib/python3.9/site-packages/interpret/visual/_udash.py:117: FutureWarning:
Series.__getitem__ treating keys as positions is deprecated. In a future version, integer keys will always be treated as labels (consistent with DataFrame behavior). To access a value by position, use `ser.iloc[pos]`
# Getting into the guts of the EBM model - We can do a lot more. But these plots shows that the EBM also says importance is the distributoons between the label.
df = pd.DataFrame({
'names': [name.replace('_scaled', '') for name in ebm_global._internal_obj['overall']['names']],
'scores': ebm_global._internal_obj['overall']['scores']
})
# Sort the dataframe by scores from highest to lowest
df_sorted = df.sort_values(by='scores', ascending=False)
# pull 15, the same as the UI shows above.
features_to_plot = df_sorted['names'].head(15).tolist()
if(CFG.CLEARML_ON):
clearml_ebm.log_data(data=features_to_plot,title='EBM Feature Importance List')
## Lets test our stats.
plot_feature_distribution(df_essays_copy,features_to_plot, True)
from torch.nn.functional import softmax
essays_for_inference_final = pd.read_csv('/Users/lange/dev/ai-or-biology/data/train_drcat_04.csv')
#essays_for_inference_final = pd.read_csv('/kaggle/input/llm-detect-ai-generated-text/test_essays.csv')
essays_for_inference_1 = essays_for_inference_final[essays_for_inference_final['label'] == 1].sample(20)
essays_for_inference_0 = essays_for_inference_final[essays_for_inference_final['label'] == 0].sample(20) #essays_for_inference[essays_for_inference['label'] == 0].sample(len(essays_for_inference_train))
essays_for_inference_cntat = pd.concat([essays_for_inference_1, essays_for_inference_0])
essays_for_inference_final = essays_for_inference_cntat.sample(frac=1).reset_index(drop=True)
import joblib
from sklearn.preprocessing import MinMaxScaler, StandardScaler
import pandas as pd
import traceback
def scale_columns(df, columns_to_scale, scaler=None, scale_type='MinMaxScaler'):
"""
Scale the specified columns in a DataFrame and add a suffix to the column names.
Args:
df (pandas.DataFrame): The DataFrame to scale.
columns_to_scale (list): List of column names to scale.
scaler (object, optional): Scaler object to use for scaling. If None, a new scaler object will be created.
scale_type (str, optional): The type of scaler to use. Default is 'MinMaxScaler'. Options: 'MinMaxScaler', 'StandardScaler'.
Returns:
pandas.DataFrame: The full DataFrame with scaled columns added.
pandas.DataFrame: A separate DataFrame with only the specified columns scaled.
object: The scaler object used for scaling.
"""
if scale_type == 'MinMaxScaler':
scaler = MinMaxScaler() if scaler is None else scaler
elif scale_type == 'StandardScaler':
scaler = StandardScaler() if scaler is None else scaler
else:
raise ValueError("Invalid scale_type. Options: 'MinMaxScaler', 'StandardScaler'")
scaled_columns = scaler.transform(df[columns_to_scale])
scaled_df = pd.DataFrame(scaled_columns, columns=[col + '_scaled' for col in columns_to_scale])
full_df = pd.concat([df.drop(columns=columns_to_scale), scaled_df], axis=1)
return full_df, scaled_df, scaler
def generate_features_for_inference(test_essays):
try:
#df_essays_copy = pd.read_pickle("scratch/df_essays_copy.pkl")
## Run them through the pipeline to get the features
df_essays = pipeline_preprocess_text(test_essays)
df_essays = process_readability_scores(df_essays)
df_essays = process_semantic_density(df_essays)
df_essays = process_semantic_flow_variability(df_essays)
df_essays = apply_empath_analysis(df_essays)
df_essays["textual_entropy"] = df_essays["text"].progress_apply(calculate_entropy)
df_essays = process_syntactic_tree_patterns(df_essays)
readability_columns = ['flesch_kincaid_grade', 'gunning_fog', 'coleman_liau_index', 'smog_index', 'ari', 'dale_chall', 'textual_entropy', 'semantic_density', 'semantic_flow_variability']
scaler = joblib.load(f"{CFG.SCRATCH_PATH}/scaler_semantic_features.pkl")
# Scale the columns using MinMaxScaler
readability_scaled_backin_df, readability_scaled_df, readability_scaler = scale_columns(df_essays, readability_columns,scaler, scale_type='MinMaxScaler')
psycho_columns = ['help','office','dance','money','wedding','domestic_work','sleep','medical_emergency','cold','hate','cheerfulness','aggression','occupation','envy','anticipation','family','vacation','crime','attractive','masculine','prison','health','pride','dispute','nervousness','government','weakness','horror','swearing_terms','leisure','suffering','royalty','wealthy','tourism','furniture','school','magic','beach','journalism','morning','banking','social_media','exercise','night','kill','blue_collar_job','art','ridicule','play','computer','college','optimism','stealing','real_estate','home','divine','sexual','fear','irritability','superhero','business','driving','pet','childish','cooking','exasperation','religion','hipster','internet','surprise','reading','worship','leader','independence','movement','body','noise','eating','medieval','zest','confusion','water','sports','death','healing','legend','heroic','celebration','restaurant','violence','programming','dominant_heirarchical','military','neglect','swimming','exotic','love','hiking','communication','hearing','order','sympathy','hygiene','weather','anonymity','trust','ancient','deception','fabric','air_travel','fight','dominant_personality','music','vehicle','politeness','toy','farming','meeting','war','speaking','listen','urban','shopping','disgust','fire','tool','phone','gain','sound','injury','sailing','rage','science','work','appearance','valuable','warmth','youth','sadness','fun','emotional','joy','affection','traveling','fashion','ugliness','lust','shame','torment','economics','anger','politics','ship','clothing','car','strength','technology','breaking','shape_and_size','power','white_collar_job','animal','party','terrorism','smell','disappointment','poor','plant','pain','beauty','timidity','philosophy','negotiate','negative_emotion','cleaning','messaging','competing','law','friends','payment','achievement','alcohol','liquid','feminine','weapon','children','monster','ocean','giving','contentment','writing','rural','positive_emotion','musical']
# Scale the columns using MinMaxScaler
scaler_psyco = joblib.load(f"{CFG.SCRATCH_PATH}/scaler_psycho_features.pkl")
psycho_scaled_df_backin_df, psycho_scaled_df, psycho_scaler = scale_columns(df_essays, psycho_columns,scaler_psyco, scale_type='MinMaxScaler')
# Define the columns to scale
text_features = ['num_sentences', 'num_tokens', 'num_unique_lemmas', 'average_token_length', 'average_sentence_length', 'num_entities', 'num_noun_chunks', 'num_pos_tags', 'num_distinct_entities', 'average_entity_length', 'average_noun_chunk_length', 'max_depth', 'avg_branching_factor', 'total_nodes', 'total_leaves', 'unique_rules', 'tree_complexity', 'depth_variability']
scaler_text = joblib.load(f"{CFG.SCRATCH_PATH}/scaler_tree_features.pkl")
# Scale the columns using MinMaxScaler
tree_feature_scaler_backin_df, tree_features_scaled_df, tree_feature_scaler = scale_columns(df_essays,
text_features,scaler_text, scale_type='MinMaxScaler')
final_features_df = pd.concat([readability_scaled_df,tree_features_scaled_df,psycho_scaled_df], axis=1)
#print(final_features_df.head())
return final_features_df
except Exception as e:
# if any fail, revert to the bert inference
print(f"Error in feature extraction: {e}")
import torch
import pandas as pd
def bert_inference(dataframe, model, tokenizer, max_length, device):
"""
Performs inference on a dataframe using a pre-loaded model and returns softmax probabilities.
Args:
- dataframe (pd.DataFrame): DataFrame containing the texts to classify.
- model (torch.nn.Module): Pre-loaded trained model for inference.
- tokenizer (transformers.PreTrainedTokenizer): Tokenizer for the model.
- max_length (int): Maximum sequence length for tokenization.
- device (torch.device): The device to run the model on (CPU or GPU).
Returns:
- pd.DataFrame: Original DataFrame with additional columns for predictions and probabilities.
"""
model.to(device)
model.eval()
predictions = []
probabilities = []
for _, row in dataframe.iterrows():
text = row['text']
inputs = tokenizer.encode_plus(
text,
add_special_tokens=True,
max_length=max_length,
padding='max_length',
truncation=True,
return_tensors='pt'
)
input_ids = inputs['input_ids'].to(device)
attention_mask = inputs['attention_mask'].to(device)
with torch.no_grad():
output = model(input_ids, attention_mask)
logits = output.logits if hasattr(output, 'logits') else output
probs = torch.nn.functional.softmax(logits, dim=1)
probabilities_ind = torch.nn.functional.softmax(probs, dim=1).cpu().numpy()[0]
prediction = torch.argmax(probs, dim=1).cpu().numpy()[0]
if (prediction>= 1):
probability = max(probabilities_ind)
else:
probability = min(probabilities_ind)
predictions.append(prediction)
probabilities.append(probability)
dataframe['predicted_label'] = predictions
dataframe['probability'] = probabilities
return dataframe
import pandas as pd
import numpy as np
from scipy.special import softmax
def ebm_inference(model, data):
"""
Performs inference using an EBM model and returns predictions with probabilities.
Args:
- model: Trained EBM model.
- data (pd.DataFrame): DataFrame containing the data for inference.
Returns:
- pd.DataFrame: DataFrame with predictions and corresponding probabilities.
"""
X = data.values
# Initialize lists for predictions and probabilities
predictions = []
probabilities = []
# Iterate over each row in the input data
for i in range(len(X)):
# Get the current row and reshape it
row = X[i].reshape(1, -1)
# Make prediction and calculate probabilities
prediction = model.predict(row)
probabilities_ind = model.predict_proba(row)
softmax_probabilities = softmax(probabilities_ind, axis=1) # ebm prob is a sigmoid, we need softmax which extends sigmoid
# Append the prediction and probability to the lists
predictions.append(prediction[0])
probabilities.append(softmax_probabilities[0][1])
# Combine predictions and probabilities into a single DataFrame
results = pd.DataFrame({
'predicted_label': predictions,
'probability': probabilities
})
return results
df_to_classify = pipeline_preprocess_text(essays_for_inference_final)
100%|██████████| 40/40 [00:00<00:00, 205.28it/s]
Preprocessing completed in 0.20 seconds
from transformers import BertTokenizer, BertForSequenceClassification, TrainingArguments
import torch
from torch import nn
from transformers import BertModel
#if(CFG.EBM_ONLY == False):
import torch
from torchsummary import summary
# This is sloppy - but leaving for now asit works. Will clean up later.
# Configuration and Usage
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# Load the tokenizer, study, and best model
import pickle
with open(f'{CFG.SCRATCH_PATH}/bert_seq_class_tokeniser.pkl', 'rb') as f:
tokenizer = pickle.load(f)
with open(f'{CFG.SCRATCH_PATH}/bert_seq_class_study.pkl', 'rb') as f:
study = pickle.load(f)
model_config = {
'bert_model': CFG.BERT_MODEL,
'do_lower_case': False,
'num_labels': 2,
'output_attentions': False,
'output_hidden_states': False,
'max_length': 32,
'optuna_trials': 1,
'epochs': 1,
'device': torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')
}
# Copy of the args from the model. putting them here just in case it run wothout that cell.
training_args = TrainingArguments(
output_dir=f'{CFG.SCRATCH_PATH}/results',
num_train_epochs=model_config['epochs'],
per_device_train_batch_size=16,
per_device_eval_batch_size=32,
warmup_steps=100,
weight_decay=0.01,
logging_dir=f'{CFG.SCRATCH_PATH}/logs/bert_sequence_classifier',
learning_rate=5e-5,
adam_epsilon=1e-8,
do_train=True,
do_eval=True,
no_cuda=False,
evaluation_strategy="epoch",
save_strategy="steps",
logging_steps=500,
save_steps=500,
)
# Load the model
## Get params from the best trial
#best_trial_params = study.best_trial.params
# set the training arguments with the best trail values
training_args.learning_rate = study["learning_rate"]
training_args.per_device_train_batch_size = study["batch_size"]
# load the model with the best OPTUNA trial
best_model = BertClassifier(training_args, model_config)
#best_model.model.load_state_dict(torch.load(f"{CFG.SCRATCH_PATH}/bert_sequence_classifier_best.pt"))
best_model.model.load_state_dict(torch.load(f"{CFG.SCRATCH_PATH}/bert_sequence_classifier_best.pt",map_location=device))
print("----- Bert For Sequence Classification -----")
print(best_model.model)
# Run Inference
bert_seq_classifier_inference_results = bert_inference(df_to_classify, best_model.model, tokenizer,
model_config['max_length'], device)
if(CFG.KAGGLE_RUN == False):
correct_predictions = (bert_seq_classifier_inference_results['label'] == bert_seq_classifier_inference_results['predicted_label']).sum()
# Calculate the total number of predictions
total_predictions = len(bert_seq_classifier_inference_results)
# Calculate the accuracy
accuracy = correct_predictions / total_predictions
# Print the statistics
print(f"Total predictions: {total_predictions}")
print(f"Correct predictions: {correct_predictions}")
print(f"Accuracy: {accuracy}")
# Get the true labels and predicted labels as numpy arrays
true_labels = bert_seq_classifier_inference_results['label'].to_numpy()
predicted_labels = bert_seq_classifier_inference_results['predicted_label'].to_numpy()
# Calculate the confusion matrix
cm = confusion_matrix(true_labels, predicted_labels)
# Print the confusion matrix
print("Confusion Matrix:")
print(cm)
incorrect_row_numbers = np.where(true_labels != predicted_labels)[0]
# Print the row numbers of incorrect predictions
print("Incorrect Row Numbers:")
print(incorrect_row_numbers)
Some weights of BertForSequenceClassification were not initialized from the model checkpoint at bert-base-uncased and are newly initialized: ['classifier.bias', 'classifier.weight']
You should probably TRAIN this model on a down-stream task to be able to use it for predictions and inference.
----- Bert For Sequence Classification -----
BertForSequenceClassification(
(bert): BertModel(
(embeddings): BertEmbeddings(
(word_embeddings): Embedding(30522, 768, padding_idx=0)
(position_embeddings): Embedding(512, 768)
(token_type_embeddings): Embedding(2, 768)
(LayerNorm): LayerNorm((768,), eps=1e-12, elementwise_affine=True)
(dropout): Dropout(p=0.1, inplace=False)
)
(encoder): BertEncoder(
(layer): ModuleList(
(0-11): 12 x BertLayer(
(attention): BertAttention(
(self): BertSelfAttention(
(query): Linear(in_features=768, out_features=768, bias=True)
(key): Linear(in_features=768, out_features=768, bias=True)
(value): Linear(in_features=768, out_features=768, bias=True)
(dropout): Dropout(p=0.1, inplace=False)
)
(output): BertSelfOutput(
(dense): Linear(in_features=768, out_features=768, bias=True)
(LayerNorm): LayerNorm((768,), eps=1e-12, elementwise_affine=True)
(dropout): Dropout(p=0.1, inplace=False)
)
)
(intermediate): BertIntermediate(
(dense): Linear(in_features=768, out_features=3072, bias=True)
(intermediate_act_fn): GELUActivation()
)
(output): BertOutput(
(dense): Linear(in_features=3072, out_features=768, bias=True)
(LayerNorm): LayerNorm((768,), eps=1e-12, elementwise_affine=True)
(dropout): Dropout(p=0.1, inplace=False)
)
)
)
)
(pooler): BertPooler(
(dense): Linear(in_features=768, out_features=768, bias=True)
(activation): Tanh()
)
)
(dropout): Dropout(p=0.1, inplace=False)
(classifier): Linear(in_features=768, out_features=2, bias=True)
)
Total predictions: 40
Correct predictions: 38
Accuracy: 0.95
Confusion Matrix:
[[20 0]
[ 2 18]]
Incorrect Row Numbers:
[ 1 24]
model_config = {
'bert_model_name': CFG.BERT_MODEL,
'num_classes': 2,
'max_length': 128,
'batch_size': 16,
'num_epochs': 4,
'train_data': df_essays_copy,
'num_trials': 2,
}
# Pickle the tokenizer, study, and best model
with open(f'{CFG.SCRATCH_PATH}/custom_bert_tokenizer.pkl', 'rb') as f:
tokenizer = pickle.load(f)
with open(f'{CFG.SCRATCH_PATH}/best_custom_model_study.pkl', 'rb') as f:
study = pickle.load(f)
# Retrain model with best hyperparameters
best_trial = study.best_trial
#Load the model with the best trial
best_trial_params = study.best_trial.params
learning_rate = best_trial_params["learning_rate"]
dropout_rate = best_trial_params["dropout_rate"]
fc_layer_size = best_trial_params["fc_layer_size"]
lstm_hidden_size = best_trial_params["lstm_hidden_size"]
lstm_layers = best_trial_params["lstm_layers"]
best_model = BERTBiLSTMClassifier(model_config['bert_model_name'], model_config['num_classes'], dropout_rate, fc_layer_size,lstm_layers)
best_model.load_state_dict(torch.load(f'{CFG.SCRATCH_PATH}/bert_finetune_custom_best.pt',map_location=device))
#Initialize the best model with the optimal hyperparameters
best_model.to(device)
print(best_model)
custom_bert_inference_results = bert_inference(df_to_classify, best_model, tokenizer, model_config['max_length'], device)
if(CFG.KAGGLE_RUN == False):
## STATISTICS
#Calculate the number of correct predictions
correct_predictions = (custom_bert_inference_results['label'] == custom_bert_inference_results['predicted_label']).sum()
# Calculate the total number of predictions
total_predictions = len(custom_bert_inference_results)
# Calculate the accuracy
accuracy = correct_predictions / total_predictions
# Print the statistics
print(f"Total predictions: {total_predictions}")
print(f"Correct predictions: {correct_predictions}")
print(f"Accuracy: {accuracy}")
# Get the true labels and predicted labels as numpy arrays
true_labels = custom_bert_inference_results['label'].to_numpy()
predicted_labels = custom_bert_inference_results['predicted_label'].to_numpy()
# Calculate the confusion matrix
cm = confusion_matrix(true_labels, predicted_labels)
if(CFG.CLEARML_ON):
clearml_bertmodel_custom.log_data(data=cm,title='Custom Bert Classifier Inference Results Confusoon Matric')
# Print the confusion matrix
print("Confusion Matrix:")
print(cm)
incorrect_row_numbers = np.where(true_labels != predicted_labels)[0]
# Print the row numbers of incorrect predictions
print("Incorrect Row Numbers:")
print(incorrect_row_numbers)
BERTBiLSTMClassifier(
(bert): BertModel(
(embeddings): BertEmbeddings(
(word_embeddings): Embedding(30522, 768, padding_idx=0)
(position_embeddings): Embedding(512, 768)
(token_type_embeddings): Embedding(2, 768)
(LayerNorm): LayerNorm((768,), eps=1e-12, elementwise_affine=True)
(dropout): Dropout(p=0.1, inplace=False)
)
(encoder): BertEncoder(
(layer): ModuleList(
(0-11): 12 x BertLayer(
(attention): BertAttention(
(self): BertSelfAttention(
(query): Linear(in_features=768, out_features=768, bias=True)
(key): Linear(in_features=768, out_features=768, bias=True)
(value): Linear(in_features=768, out_features=768, bias=True)
(dropout): Dropout(p=0.1, inplace=False)
)
(output): BertSelfOutput(
(dense): Linear(in_features=768, out_features=768, bias=True)
(LayerNorm): LayerNorm((768,), eps=1e-12, elementwise_affine=True)
(dropout): Dropout(p=0.1, inplace=False)
)
)
(intermediate): BertIntermediate(
(dense): Linear(in_features=768, out_features=3072, bias=True)
(intermediate_act_fn): GELUActivation()
)
(output): BertOutput(
(dense): Linear(in_features=3072, out_features=768, bias=True)
(LayerNorm): LayerNorm((768,), eps=1e-12, elementwise_affine=True)
(dropout): Dropout(p=0.1, inplace=False)
)
)
)
)
(pooler): BertPooler(
(dense): Linear(in_features=768, out_features=768, bias=True)
(activation): Tanh()
)
)
(lstm): LSTM(768, 32, num_layers=3, batch_first=True, bidirectional=True)
(dropout): Dropout(p=0.0456841451290286, inplace=False)
(fc): Linear(in_features=64, out_features=2, bias=True)
(relu): ReLU()
)
Total predictions: 40
Correct predictions: 39
Accuracy: 0.975
Confusion Matrix:
[[20 0]
[ 1 19]]
Incorrect Row Numbers:
[1]
import pandas as pd
import numpy as np
from interpret.glassbox import ExplainableBoostingClassifier
from scipy.special import softmax
# This creates all the features for each text field in the dataframe.
essays_for_inference_sample = generate_features_for_inference(df_to_classify)
# create our features df with the columns named correctly
features = pd.DataFrame(essays_for_inference_sample, columns=essays_for_inference_sample.columns)
# Set logging level to ERROR
logging.getLogger('interpret').setLevel(logging.ERROR)
# with open(f'{CFG.SCRATCH_PATH}/ebm.pkl', 'rb') as f:
# ebm = ebm.load(f)
## Get yout goods and bads
ebm_inference_results = ebm_inference(ebm_gs, features)
if(CFG.CLEARML_ON):
clearml_ebm.log_data(data=features,title='Explainable Boosting Model Inference Data')
clearml_ebm.log_data(data=ebm_inference_results,title='Explainable Boosting Model Inference Results')
## Output the model stats
if(CFG.KAGGLE_RUN == False):
labels_df = df_to_classify['label']
correct_predictions = (df_to_classify['label'].reset_index(drop=True) == ebm_inference_results['predicted_label'].reset_index(drop=True)).sum()
# Calculate the total number of predictions
total_predictions = len(ebm_inference_results)
# Calculate the accuracy
accuracy = correct_predictions / total_predictions
# Print the statistics
print(f"Total predictions: {total_predictions}")
print(f"Correct predictions: {correct_predictions}")
print(f"Accuracy: {accuracy}")
# Get the true labels and predicted labels as numpy arrays
true_labels = df_to_classify['label'].to_numpy()
predicted_labels = ebm_inference_results['predicted_label'].to_numpy()
# Calculate the confusion matrix
cm = confusion_matrix(true_labels, predicted_labels)
# Print the confusion matrix
print("Confusion Matrix:")
print(cm)
incorrect_row_numbers = np.where(true_labels != predicted_labels)[0]
# Print the row numbers of incorrect predictions
print("Incorrect Row Numbers:")
print(incorrect_row_numbers)
100%|██████████| 40/40 [00:00<00:00, 247.12it/s]
Preprocessing completed in 0.16 seconds
Total predictions: 40
Correct predictions: 35
Accuracy: 0.875
Confusion Matrix:
[[18 2]
[ 3 17]]
Incorrect Row Numbers:
[ 1 11 27 29 36]
if(CFG.KAGGLE_RUN == False):
from interpret.perf import ROC
ebm_perf = ROC(ebm.predict_proba).explain_perf(X_test, y_test, name='EBM')
show(ebm_perf)
if(CFG.ENSAMBLE_STRATEGY == 1): # For three models
majority_label = (bert_seq_classifier_inference_results['predicted_label'] +
custom_bert_inference_results['predicted_label'] +
ebm_inference_results['predicted_label']).apply(lambda x: 1 if x >= 1 else 0)
average_probability = (bert_seq_classifier_inference_results['probability'] +
custom_bert_inference_results['probability'] +
ebm_inference_results['probability']) / 3
if(CFG.ENSAMBLE_STRATEGY == 2): # For 2 models - best one
majority_label = (#bert_seq_classifier_inference_results['predicted_label'] +
custom_bert_inference_results['predicted_label'] +
ebm_inference_results['predicted_label']).apply(lambda x: 1 if x >= 1 else 0)
average_probability = (#bert_seq_classifier_inference_results['probability'] +
custom_bert_inference_results['probability'] +
ebm_inference_results['probability']) / 2
# Final Decision: If labels agree, use them. If not, use average probability.
# Ensamble AVG strat 1
# =================
# CFG.ENSAMBLE_STRATEGY 1
if(CFG.ENSAMBLE_STRATEGY == 1):
final_decision = majority_label.copy()
for i in range(len(bert_seq_classifier_inference_results)):
if ebm_inference_results['predicted_label'][i] != custom_bert_inference_results['predicted_label'][i]: # Best Models Disagree
final_decision[i] = 1 if average_probability[i] >= 0.50 else 0
# =================
# CFG.ENSAMBLE_STRATEGY 2
if(CFG.ENSAMBLE_STRATEGY == 2):
final_decision = majority_label.copy()
for i in range(len(bert_seq_classifier_inference_results)):
if ebm_inference_results['predicted_label'][i] != custom_bert_inference_results['predicted_label'][i]: # Best Models Disagree
final_decision[i] = 1 if average_probability[i] >= 0.50 else 0
ensemble_results = pd.DataFrame({
'final_predicted_label': final_decision,
#'label': custom_bert_inference_results['label'],
'average_probability': average_probability,
#'source': custom_bert_inference_results['source'],
'text': custom_bert_inference_results['text']
})
if(CFG.CLEARML_ON):
clearml_ebm.log_data(data=ensemble_results,title='Exsemble Model Scoring Results')
if(CFG.KAGGLE_RUN == False):
## Ensamble ------------------------
correct_predictions = (df_to_classify['label'].reset_index(drop=True) ==
ensemble_results['final_predicted_label'].reset_index(drop=True)).sum()
# Calculate the total number of predictions
total_predictions = len(ensemble_results)
# Calculate the accuracy
accuracy = correct_predictions / total_predictions
# Print the statistics
print(f"Total predictions: {total_predictions}")
print(f"Correct predictions: {correct_predictions}")
print(f"Accuracy: {accuracy}")
# Get the true labels and predicted labels as numpy arrays
true_labels = df_to_classify['label'].to_numpy()
predicted_labels = ensemble_results['final_predicted_label'].to_numpy()
predicted_averages = ensemble_results['average_probability'].to_numpy()
# Calculate the confusion matrix
cm = confusion_matrix(true_labels, predicted_labels)
if(CFG.CLEARML_ON):
clearml_ebm.log_data(data=cm,title='Exsemble Model Scoring Results Confusoon Matric')
# Print the confusion matrix
print("Confusion Matrix:")
print(cm)
incorrect_row_numbers = np.where(true_labels != predicted_labels)[0]
# Print the row numbers of incorrect predictions
print("Incorrect Row Numbers:")
print(incorrect_row_numbers)
print(predicted_averages)
Total predictions: 40
Correct predictions: 38
Accuracy: 0.95
Confusion Matrix:
[[19 1]
[ 1 19]]
Incorrect Row Numbers:
[ 1 36]
[0.69439393 0.33834966 0.70149138 0.7054628 0.3089722 0.70524129
0.31124632 0.308972 0.31014631 0.30891109 0.65973125 0.4364831
0.31098962 0.7054017 0.31575136 0.69931882 0.3998628 0.69748983
0.70319676 0.30904756 0.30995664 0.3093674 0.31151034 0.7049542
0.70468167 0.68598632 0.70542308 0.51682635 0.30910718 0.58167151
0.30929284 0.30888784 0.70514635 0.69362241 0.30898159 0.311092
0.5090657 0.70544873 0.70528596 0.30920638]
# print(bert_seq_classifier_inference_results['probability'])
# print(custom_bert_inference_results['probability'])
# print(ebm_inference_results['probability'])
🎙 References
Relating Natural Language Aptitude to Individual Differences in Learning Programming Languages) [1]
@article{article,
author = {Prat, Chantel and Madhyastha, Tara and Mottarella, Malayka and Kuo, Chu-Hsuan},
year = {2020},
month = {03},
pages = {},
title = {Relating Natural Language Aptitude to Individual Differences in Learning Programming Languages},
volume = {10},
journal = {Scientific Reports},
doi = {10.1038/s41598-020-60661-8}
}
InterpretML: A Unified Framework for Machine Learning Interpretability" (H. Nori, S. Jenkins, P. Koch, and R. Caruana 2019)[2]
@article{nori2019interpretml,
title={InterpretML: A Unified Framework for Machine Learning Interpretability},
author={Nori, Harsha and Jenkins, Samuel and Koch, Paul and Caruana, Rich},
journal={arXiv preprint arXiv:1909.09223},
year={2019}
}
The Psychological Meaning of Words: LIWC and Computerized Text Analysis Methods[3]
@article{article,
author = {Tausczik, Yla and Pennebaker, James},
year = {2010},
month = {03},
pages = {24-54},
title = {The Psychological Meaning of Words: LIWC and Computerized Text Analysis Methods},
volume = {29},
journal = {Journal of Language and Social Psychology},
doi = {10.1177/0261927X09351676}
}
“Attention is all you need” (Vaswani, Ashish & Shazeer, Noam & Parmar, Niki & Uszkoreit, Jakob & Jones, Llion & Gomez, Aidan & Kaiser, Lukasz & Polosukhin, Illia. (2017)) [4]
@misc{vaswani2023attention,
title={Attention Is All You Need},
author={Ashish Vaswani and Noam Shazeer and Niki Parmar and Jakob Uszkoreit and Llion Jones and Aidan N. Gomez and Lukasz Kaiser and Illia Polosukhin},
year={2023},
eprint={1706.03762},
archivePrefix={arXiv},
primaryClass={cs.CL}
}
Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics[5]
@inproceedings{kitaev-etal-2019-multilingual,
title = "Multilingual Constituency Parsing with Self-Attention and Pre-Training",
author = "Kitaev, Nikita and
Cao, Steven and
Klein, Dan",
booktitle = "Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics",
month = jul,
year = "2019",
address = "Florence, Italy",
publisher = "Association for Computational Linguistics",
url = "https://www.aclweb.org/anthology/P19-1340",
doi = "10.18653/v1/P19-1340",
pages = "3499--3505",
}
Constituency Parsing with a Self-Attentive Encoder[6]
@inproceedings{kitaev-klein-2018-constituency,
title = "Constituency Parsing with a Self-Attentive Encoder",
author = "Kitaev, Nikita and
Klein, Dan",
booktitle = "Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers)",
month = jul,
year = "2018",
address = "Melbourne, Australia",
publisher = "Association for Computational Linguistics",
url = "https://www.aclweb.org/anthology/P18-1249",
doi = "10.18653/v1/P18-1249",
pages = "2676--2686",
}
# # # End the timer
# # end_time = time.time()
# # # Calculate the total execution time
# # total_time = end_time - start_time
# print(f"Total execution time: {total_time} seconds")
# print(f"Total execution time: {total_time/60} minues")
# print(f"Total execution time: {total_time/60/60} hours")
# ## Stats - source GPT4
# # # End the timer
# # end_time = time.time()
# # # Calculate the total execution time
# # total_time = end_time - start_time
# import psutil
# # Get CPU information
# #cpu_info = psutil.cpu_freq()
# cpu_percent = psutil.cpu_percent()
# # Get memory information
# memory_info = psutil.virtual_memory()
# # Get disk usage information
# disk_usage = psutil.disk_usage('/')
# # Print the hardware and software specifications
# #print("CPU Frequency:", cpu_info.current, "MHz")
# print("Total Memory:", memory_info.total // (1024 ** 3), "GB")
# print("Disk Usage:", disk_usage.used // (1024 ** 3), "GB")
# # get device
# device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# print("cuda? :", device)
# labels = df_essays_copy['label'].tolist()
# features = readability_scaled_df #psycho_scaled_df
# X_train, X_test, y_train, y_test = train_test_split(features, labels, test_size=0.2, random_state=42)
# ebm = ExplainableBoostingClassifier(interactions=0,
# feature_names=features.columns.tolist())
# ebm.fit(X_train, y_train)
# ebm.score(X_test, y_test)
# from interpret import show
# ebm_global = ebm.explain_global()
# show(ebm_global)
# ebm_local = ebm.explain_local(X_test, y_test)
# show(ebm_local)
# labels = df_essays_copy['label'].tolist()
# features = psycho_scaled_df #psycho_scaled_df
# X_train, X_test, y_train, y_test = train_test_split(features, labels, test_size=0.2, random_state=42)
# ebm = ExplainableBoostingClassifier(interactions=0,
# feature_names=features.columns.tolist())
# ebm.fit(X_train, y_train)
# ebm.score(X_test, y_test)
# from interpret import show
# ebm_global = ebm.explain_global()
# show(ebm_global)
# ebm_local = ebm.explain_local(X_test, y_test)
# show(ebm_local)
# labels = df_essays_copy['label'].tolist()
# features = tree_features_scaled_df #psycho_scaled_df
# X_train, X_test, y_train, y_test = train_test_split(features, labels, test_size=0.2, random_state=42)
# ebm = ExplainableBoostingClassifier(interactions=0,
# feature_names=features.columns.tolist())
# ebm.fit(X_train, y_train)
# ebm.score(X_test, y_test)
# from interpret import show
# ebm_global = ebm.explain_global()
# show(ebm_global)
# ebm_local = ebm.explain_local(X_test, y_test)
# show(ebm_local)
# categories_to_plot = ['average_token_length','num_pos_tags','num_distinct_entities','average_entity_length','average_noun_chunk_length','max_depth','avg_branching_factor','total_nodes','total_leaves','unique_rules','tree_complexity','depth_variability']
# if(CFG.CLEARML_ON):
# plot_categories_box(df_essays, categories_to_plot,clearml_handler.task)
# else:
# plot_categories_box(df_essays, categories_to_plot)